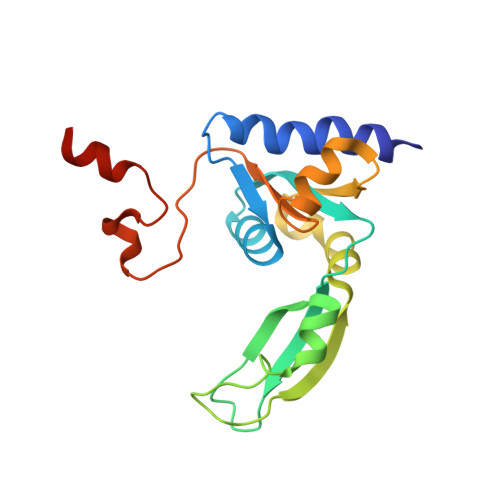
NMR solution structure and function of the C-terminal domain of eukaryotic class 1 polypeptide chain release factor.
Mantsyzov, A.B., Ivanova, E.V., Birdsall, B., Alkalaeva, E.Z., Kryuchkova, P.N., Kelly, G., Frolova, L.Y., Polshakov, V.I.(2010) FEBS J 277: 2611-2627
- PubMed: 20553496
- DOI: https://doi.org/10.1111/j.1742-464X.2010.07672.x
- Primary Citation of Related Structures:
2KTV - PubMed Abstract:
Termination of translation in eukaryotes is triggered by two polypeptide chain release factors, eukaryotic class 1 polypeptide chain release factor (eRF1) and eukaryotic class 2 polypeptide chain release factor 3. eRF1 is a three-domain protein that interacts with eukaryotic class 2 polypeptide chain release factor 3 via its C-terminal domain (C-domain). The high-resolution NMR structure of the human C-domain (residues 277-437) has been determined in solution. The overall fold and the structure of the beta-strand core of the protein in solution are similar to those found in the crystal structure. The structure of the minidomain (residues 329-372), which was ill-defined in the crystal structure, has been determined in solution. The protein backbone dynamics, studied using (15)N-relaxation experiments, showed that the C-terminal tail 414-437 and the minidomain are the most flexible parts of the human C-domain. The minidomain exists in solution in two conformational states, slowly interconverting on the NMR timescale. Superposition of this NMR solution structure of the human C-domain onto the available crystal structure of full-length human eRF1 shows that the minidomain is close to the stop codon-recognizing N-terminal domain. Mutations in the tip of the minidomain were found to affect the stop codon specificity of the factor. The results provide new insights into the possible role of the C-domain in the process of translation termination.
- Center for Magnetic Tomography and Spectroscopy, M. V. Lomonosov Moscow State University, Russia.
Organizational Affiliation: