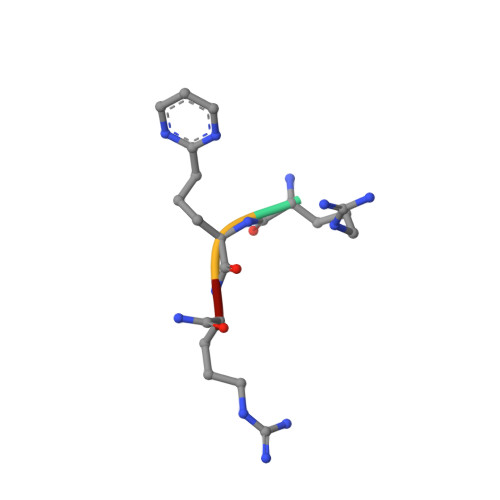
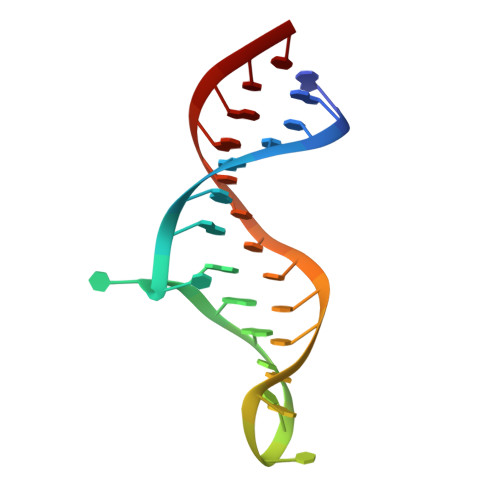
Structures of HIV TAR RNA-ligand complexes reveal higher binding stoichiometries.
Ferner, J., Suhartono, M., Breitung, S., Jonker, H.R.A., Hennig, M., Wohnert, J., Gobel, M., Schwalbe, H.(2009) Chembiochem 10: 1490-1494
- PubMed: 19444830
- DOI: https://doi.org/10.1002/cbic.200900220
- Primary Citation of Related Structures:
2KMJ - PubMed Abstract:
Target TAR by NMR: Tripeptides containing arginines as terminal residues and non-natural amino acids as central residues are good leads for drug design to target the HIV trans-activation response element (TAR). The structural characterization of the RNA-ligand complex by NMR spectroscopy reveals two specific binding sites that are located at bulge residue U23 and around the pyrimidine-stretch U40-C41-U42 directly adjacent to the bulge.
- Institut für Organische Chemie und Chemische Biologie, Zentrum für Biomolekulare Magnetische Resonanz (BMRZ), Johann Wolfgang Goethe-Universität Frankfurt am Main, Max-von-Laue-Strasse 7, 60438 Frankfurt am Main, Germany.
Organizational Affiliation: