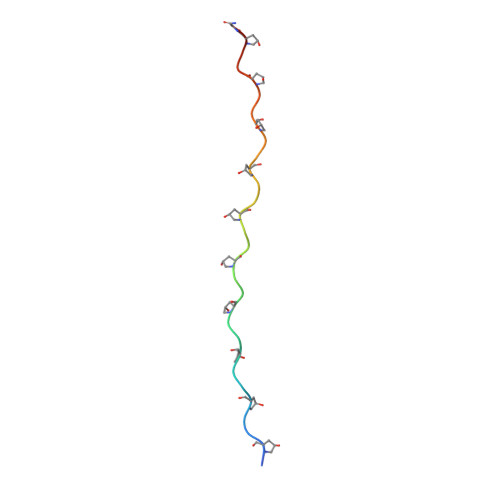
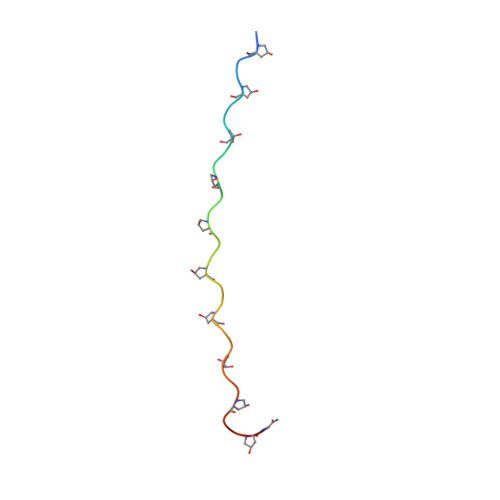
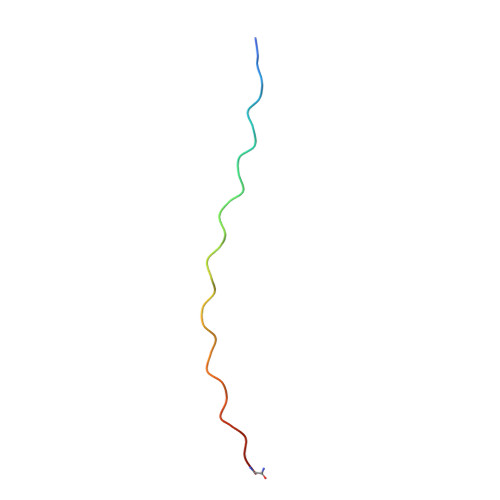Solution structure of an ABC collagen heterotrimer reveals a single-register helix stabilized by electrostatic interactions.
Fallas, J.A., Gauba, V., Hartgerink, J.D.(2009) J Biological Chem 284: 26851-26859
- PubMed: 19625247
- DOI: https://doi.org/10.1074/jbc.M109.014753
- Primary Citation of Related Structures:
2KLW - PubMed Abstract:
Collagen, known for its structural role in tissues and also for its participation in the regulation of homeostatic and pathological processes in mammals, is assembled from triple helices that can be either homotrimers or heterotrimers. High resolution structural information for natural collagens has been difficult to obtain because of their size and the heterogeneity of their native environment. For this reason, peptides that self-assemble into collagen-like triple helices are used to gain insight into the structure, stability, and biochemistry of this important protein family. Although many of the most common collagens in humans are heterotrimers, almost all studies of collagen helices have been on homotrimers. Here we report the first structure of a collagen heterotrimer. Our structure, obtained by solution NMR, highlights the role of electrostatic interactions as stabilizing factors within the triple helical folding motif. This addresses an issue that has been actively researched because of the predominance of charged residues in the collagen family. We also find that it is possible to selectively form a collagen heterotrimer with a well defined composition and register of the peptide chains within the helix, based on information encoded solely in the collagenous domain. Globular domains are implicated in determining the composition of several collagen types, but it is unclear what their role in controlling register may be. We show that is possible to design peptides that not only selectively choose a composition but also a specific register without the assistance of other protein constructs. This mechanism may be used in nature as well.
- Department of Chemistry and Bioengineering, Rice University, Houston, Texas 77005, USA.
Organizational Affiliation: