Solution NMR structure of the ARID domain of human AT-rich interactive domain-containing protein 3A: a human cancer protein interaction network target.
Liu, G., Huang, Y.J., Xiao, R., Wang, D., Acton, T.B., Montelione, G.T.(2010) Proteins 78: 2170-2175
- PubMed: 20455271
- DOI: https://doi.org/10.1002/prot.22718
- Primary Citation of Related Structures:
2KK0 - PubMed Abstract:
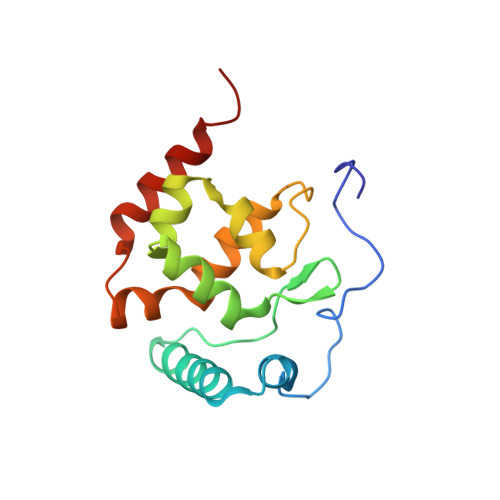
The AT-rich interactive domain (ARID) of human AT-rich interactive domain-containing protein 3A (ARID3A) has been selected for structural characterization by Northeast Structural Genomics Consortium (residues 218-351 NESG ID HR4394C) as part of our Human Cancer Protein Interaction Network (HCPIN) project. Protein ARID3A belongs to the ARID family DNA-binding protein and is known to play important roles in embryonic patterning, cell lineage gene regulation, and cell cycle control, chromatin remodeling and transcriptional regulations. The solution NMR structure of ARID3A ARID domain consists of eight α-helices α0-α7 and a short β hairpin. Helix α0 and α1 form a V shape, helix α2-α4 and helix α5-α7 form two U shapes, and the V and two U shapes packed orthogonal to each other. The NMR structure of the ARID domain of human ARID3A reported here provides a structural basis for elucidating the regulatory mechanisms of ARID3A function, and the molecular mechanism of ARID3A interactions with DNA. It also has potential value in future drug discovery and design.
- Center for Advanced Biotechnology and Medicine, Department of Molecular Biology and Biochemistry, Rutgers, The State University of New Jersey, Piscataway, New Jersey 08854, USA. gliu@cabm.rutgers.edu
Organizational Affiliation: