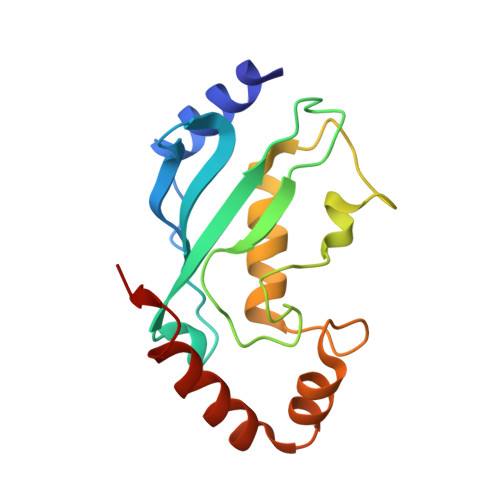
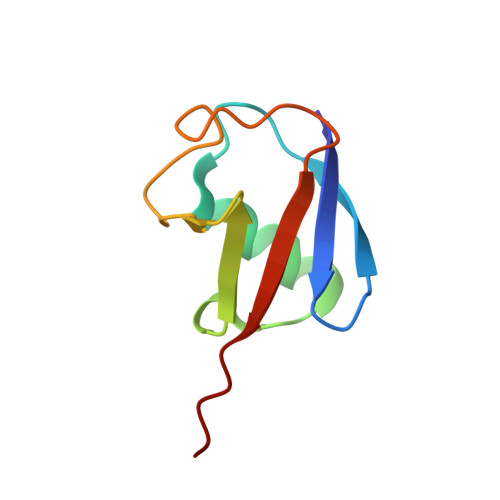
The structure of the UbcH8-ubiquitin complex shows a unique ubiquitin interaction site.
Serniwka, S.A., Shaw, G.S.(2009) Biochemistry 48: 12169-12179
- PubMed: 19928833
- DOI: https://doi.org/10.1021/bi901686j
- Primary Citation of Related Structures:
2KJH - PubMed Abstract:
Ubiquitin-mediated proteolysis utilizes a series of three key enzymes (E1, E2, and E3) to transfer and then covalently modify a substrate with ubiquitin. E2 conjugating enzymes are central proteins in this pathway responsible for the acceptance of a ubiquitin from the E1 enzyme and association with an E3 protein. All E2 enzymes covalently bind ubiquitin through a thiolester linkage between a conserved active-site cysteine on E2 and the C-terminal glycine on ubiquitin. It is not known whether E2 enzymes utilize similar surfaces and residues to coordinate a ubiquitin molecule and how this might contribute to any substrate specificity. In this work, we determined the structure of the human E2 enzyme UbcH8 (UBE2L6) covalently bound to ubiquitin by NMR spectroscopy. A disulfide bond mimicking the short-lived thiolester was formed between the two proteins providing a stable complex. Overall, the structure of UbcH8 does not undergo a significant conformational change upon forming a complex with ubiquitin. Chemical shift perturbation and cross-saturation experiments were used to identify contacts between UbcH8 and ubiquitin and those contacts used as inputs for HADDOCK molecular docking to produce the structure of the UbcH8-ubiquitin complex. An ensemble of 16 structures (root-mean-square deviation of 0.83 A) showed that ubiquitin interacts with the linker region prior to the alpha5 helix as well as residues near the catalytic site. This region corresponds to an area of negative potential on the UbcH8 surface and is considerably different from other E2-ubiquitin interaction sites. Our findings indicate the positioning of ubiquitin on UbcH8 would still allow interaction with E1 and E3 enzymes. Together, the results suggest the UbcH8-ubiquitin complex may provide an additional level of specificity in the ubiquitination pathway.
- Department of Biochemistry, University of Western Ontario, London, Ontario N6A 5C1, Canada.
Organizational Affiliation: