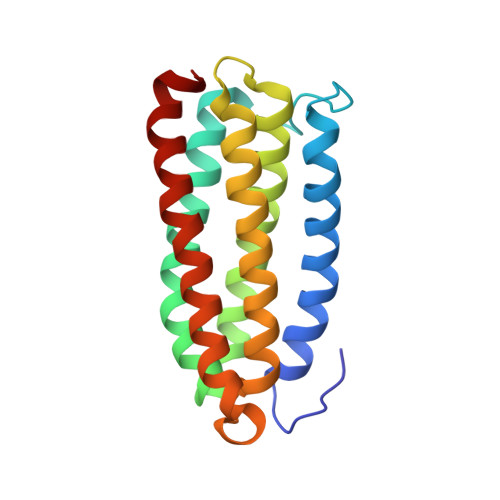
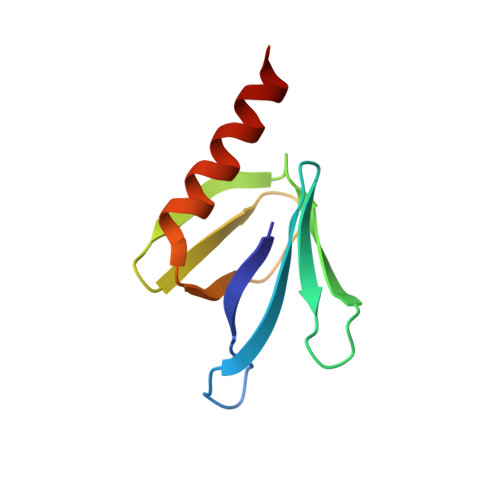
The structure of an interdomain complex that regulates talin activity.
Goult, B.T., Bate, N., Anthis, N.J., Wegener, K.L., Gingras, A.R., Patel, B., Barsukov, I.L., Campbell, I.D., Roberts, G.C., Critchley, D.R.(2009) J Biological Chem 284: 15097-15106
- PubMed: 19297334
- DOI: https://doi.org/10.1074/jbc.M900078200
- Primary Citation of Related Structures:
2KBB, 2KGX - PubMed Abstract:
Talin is a large flexible rod-shaped protein that activates the integrin family of cell adhesion molecules and couples them to cytoskeletal actin. It exists in both globular and extended conformations, and an intramolecular interaction between the N-terminal F3 FERM subdomain and the C-terminal part of the talin rod contributes to an autoinhibited form of the molecule. Here, we report the solution structure of the primary F3 binding domain within the C-terminal region of the talin rod and use intermolecular nuclear Overhauser effects to determine the structure of the complex. The rod domain (residues 1655-1822) is an amphipathic five-helix bundle; Tyr-377 of F3 docks into a hydrophobic pocket at one end of the bundle, whereas a basic loop in F3 (residues 316-326) interacts with a cluster of acidic residues in the middle of helix 4. Mutation of Glu-1770 abolishes binding. The rod domain competes with beta3-integrin tails for binding to F3, and the structure of the complex suggests that the rod is also likely to sterically inhibit binding of the FERM domain to the membrane.
- Department of Biochemistry, University of Leicester, Lancaster Road, Leicester LE1 9HN, UK.
Organizational Affiliation: