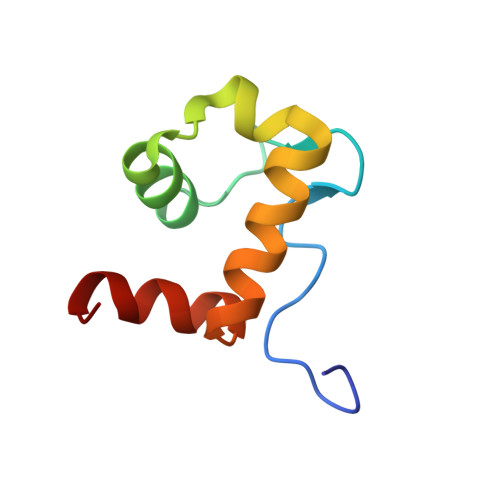
Structural mechanism of WASP activation by the enterohaemorrhagic E. coli effector EspF(U).
Cheng, H.C., Skehan, B.M., Campellone, K.G., Leong, J.M., Rosen, M.K.(2008) Nature 454: 1009-1013
- PubMed: 18650809
- DOI: https://doi.org/10.1038/nature07160
- Primary Citation of Related Structures:
2K42 - PubMed Abstract:
During infection, enterohaemorrhagic Escherichia coli (EHEC) takes over the actin cytoskeleton of eukaryotic cells by injecting the EspF(U) protein into the host cytoplasm. EspF(U) controls actin by activating members of the Wiskott-Aldrich syndrome protein (WASP) family. Here we show that EspF(U) binds to the autoinhibitory GTPase binding domain (GBD) in WASP proteins and displaces it from the activity-bearing VCA domain (for verprolin homology, central hydrophobic and acidic regions). This interaction potently activates WASP and neural (N)-WASP in vitro and induces localized actin assembly in cells. In the solution structure of the GBD-EspF(U) complex, EspF(U) forms an amphipathic helix that binds the GBD, mimicking interactions of the VCA domain in autoinhibited WASP. Thus, EspF(U) activates WASP by competing directly for the VCA binding site on the GBD. This mechanism is distinct from that used by the eukaryotic activators Cdc42 and SH2 domains, which globally destabilize the GBD fold to release the VCA. Such diversity of mechanism in WASP proteins is distinct from other multimodular systems, and may result from the intrinsically unstructured nature of the isolated GBD and VCA elements. The structural incompatibility of the GBD complexes with EspF(U) and Cdc42/SH2, plus high-affinity EspF(U) binding, enable EHEC to hijack the eukaryotic cytoskeletal machinery effectively.
- Department of Biochemistry and Howard Hughes Medical Institute, University of Texas Southwestern Medical Center, Dallas, Texas 75390, USA.
Organizational Affiliation: