Understanding the plasticity of the ubiquitin-protein recognition code: the josephin domain of ataxin-3 is a diubiquitin binding motif
Nicastro, G., Menon, R., Masino, L., Pastore, A.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ataxin-3 | 182 | Homo sapiens | Mutation(s): 0 Gene Names: ATXN3, ATX3, MJD, MJD1, SCA3 EC: 3.4.22 (PDB Primary Data), 3.4.19.12 (UniProt) | 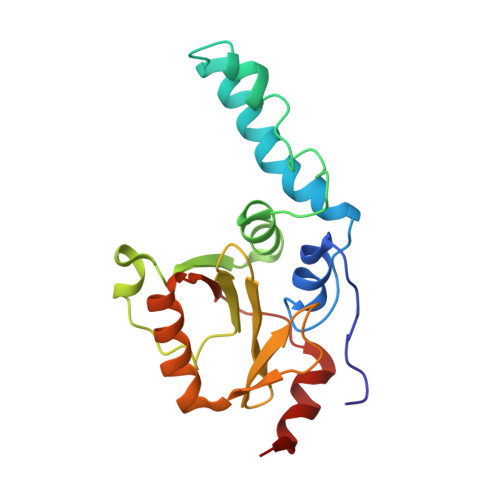 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P54252 (Homo sapiens) Explore P54252 Go to UniProtKB: P54252 | |||||
PHAROS: P54252 GTEx: ENSG00000066427 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P54252 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| UBC protein | 76 | Homo sapiens | Mutation(s): 0 Gene Names: UBC | 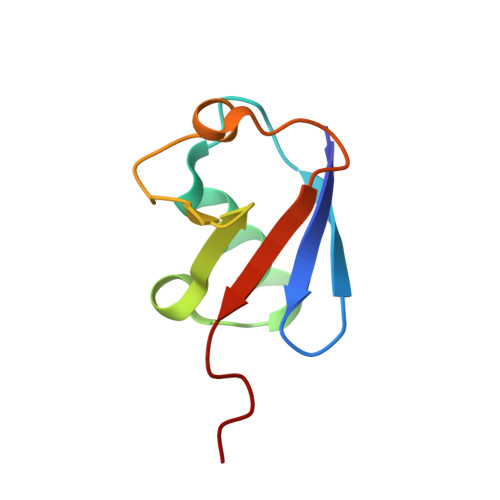 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P0CG48 (Homo sapiens) Explore P0CG48 Go to UniProtKB: P0CG48 | |||||
PHAROS: P0CG48 GTEx: ENSG00000150991 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0CG48 | ||||
Sequence AnnotationsExpand | |||||
| |||||