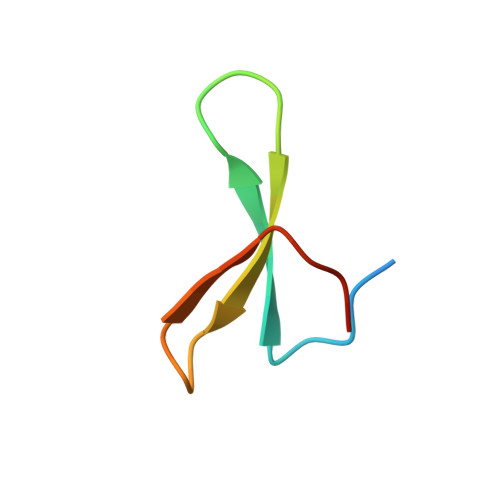
NMR Structural Studies of the ItchWW3 Domain Reveal that Phosphorylation at T30 Inhibits the Interaction with PPxY-Containing Ligands
Morales, B., Ramirez-Espain, X., Shaw, A.Z., Martin-Malpartida, P., Yraola, F., Sanchez-Tillo, E., Farrera, C., Celada, A., Royo, M., Macias, M.J.(2007) Structure 15: 473-483
- PubMed: 17437719
- DOI: https://doi.org/10.1016/j.str.2007.03.005
- Primary Citation of Related Structures:
2JO9, 2JOC - PubMed Abstract:
In this work, we study the role of phosphorylation as a regulatory mechanism for the interaction between the E3 ubiquitin ligase ItchWW3 domain and two PPxY motifs of one of its targets, the Epstein-Barr virus latent membrane protein 2A. Whereas ligand phosphorylation only diminishes binding, domain phosphorylation at residue T30 abrogates it. We show that two ItchWW domains can be phosphorylated at this position, using CK2 and PKA kinases and/or with stimulated T lymphocyte lysates. To better understand the regulation process, we determined the NMR structures of the ItchWW3-PPxY complex and of the phosphoT30-ItchWW3 variant. The peptide binds the domain using both XP and tyrosine grooves. A hydrogen bond from T30 to the ligand is also detected. This hydrogen-bond formation is precluded in the variant, explaining the inhibition upon phosphorylation. Our results suggest that phosphorylation at position 30 in ItchWW domains can be a mechanism to inhibit target recognition in vivo.
- Institute of Research in Biomedicine, Institució Catalana de Recerca i Estudis Avançats, Passeig Lluís Companys 23, 08010 Barcelona, Spain.
Organizational Affiliation: