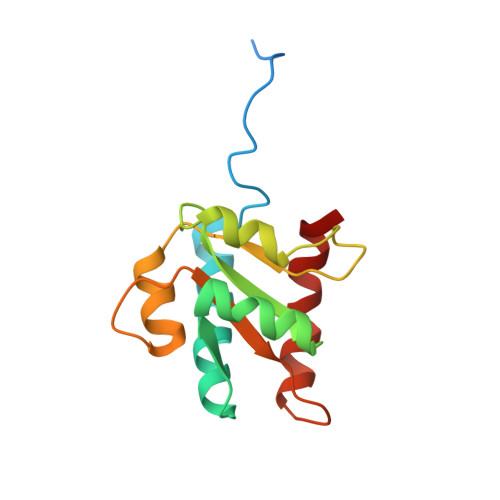
Functional Implications for a Prototypical K-Turn Binding Protein from Structural and Dynamical Studies of 15.5K
Soss, S.E., Flynn, P.F.(2007) Biochemistry 46: 14979-14986
- PubMed: 18044964
- DOI: https://doi.org/10.1021/bi701254q
- Primary Citation of Related Structures:
2JNB - PubMed Abstract:
The kink-turn (K-turn) motif is recognized and bound by a family of proteins that act as nucleation factors for ribonucleoparticle assembly. The binding of various proteins to a conserved RNA structural motif known as the K-turn has been shown to be an important component of regulation in the ribosome, in the spliceosome, and in RNA modification. 15.5K is a prototypical example of a K-turn binding protein, which has been shown to bind the 5'-U4 stem-loop of the spliceosome and the box C/D motif. We describe the solution NMR structure of free 15.5K, as well as studies of conformational flexibility from 15N NMR relaxation and H/D exchange experiments. The protein appears well-structured aside from conformational fluctuation in alpha3. Flexibility in fast time scale motions and the observation of limited intermediate and slow motions further characterize the free protein and may suggest local contributions to recognition and binding.
- Department of Chemistry, University of Utah, Salt Lake City, Utah 84112, USA.
Organizational Affiliation: