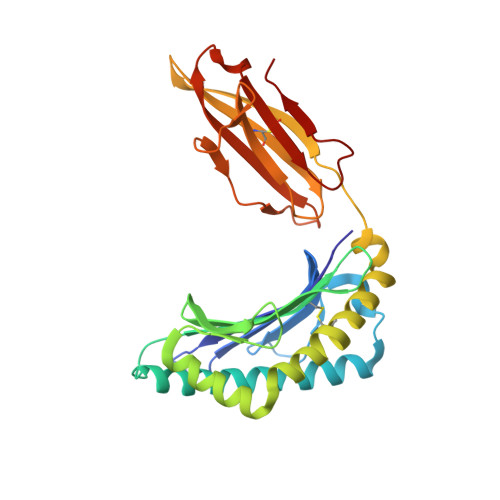
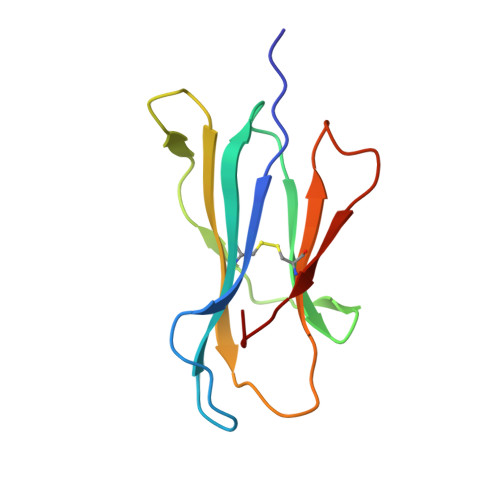
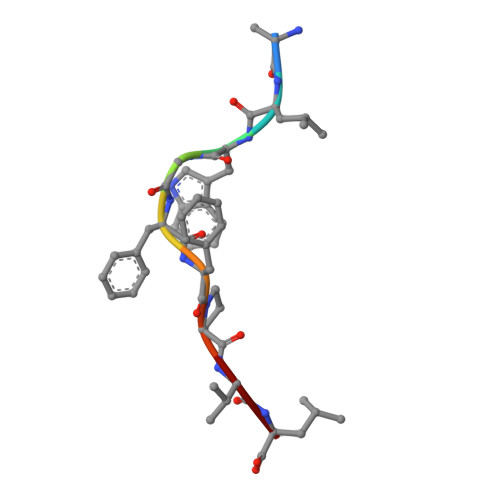
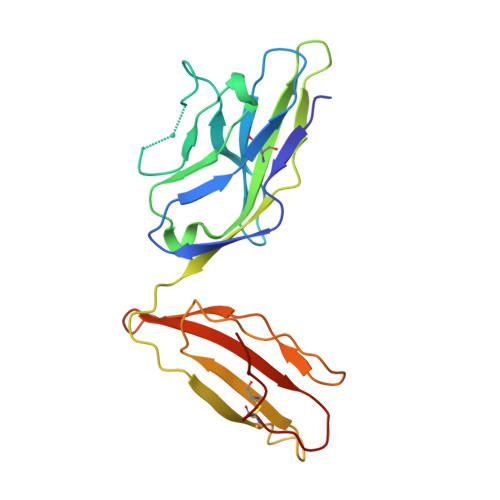
Single Mhc Mutation Eliminates Enthalpy Associated with T Cell Receptor Binding.
Miller, P., Pazy, Y., Conti, B., Riddle, D., Appella, E., Collins, E.J.(2007) J Mol Biology 373: 315
- PubMed: 17825839
- DOI: https://doi.org/10.1016/j.jmb.2007.07.028
- Primary Citation of Related Structures:
2J8U, 2JCC, 2UWE - PubMed Abstract:
The keystone of the adaptive immune response is T cell receptor (TCR) recognition of peptide presented by major histocompatibility complex (pMHC) molecules. The crystal structure of AHIII TCR bound to MHC, HLA-A2, showed a large interface with an atypical binding orientation. MHC mutations in the interface of the proteins were tested for changes in TCR recognition. From the range of responses observed, three representative HLA-A2 mutants, T163A, W167A, and K66A, were selected for further study. Binding constants and co-crystal structures of the AHIII TCR and the three mutants were determined. K66 in HLA-A2 makes contacts with both peptide and TCR, and has been identified as a critical residue for recognition by numerous TCR. The K66A mutation resulted in the lowest AHIII T cell response and the lowest binding affinity, which suggests that the T cell response may correlate with affinity. Importantly, the K66A mutation does not affect the conformation of the peptide. The change in affinity appears to be due to a loss in hydrogen bonds in the interface as a result of a conformational change in the TCR complementarity-determining region 3 (CDR3) loop. Isothermal titration calorimetry confirmed the loss of hydrogen bonding by a large loss in enthalpy. Our findings are inconsistent with the notion that the CDR1 and CDR2 loops of the TCR are responsible for MHC restriction, while the CDR3 loops interact solely with the peptide. Instead, we present here an MHC mutation that does not change the conformation of the peptide, yet results in an altered conformation of a CDR3.
- Department of Biochemistry and Biophysics, The University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA.
Organizational Affiliation: