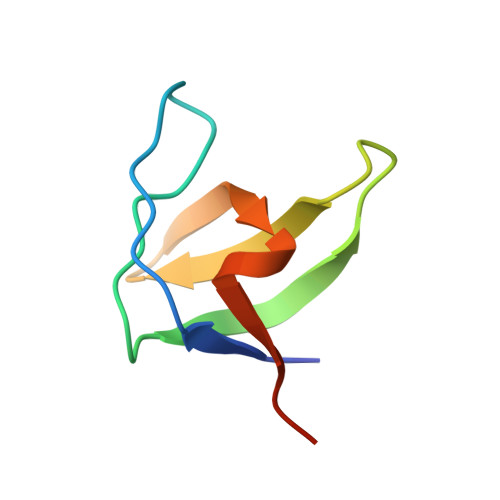
Atypical Polyproline Recognition by the Cms N-Terminal SH3 Domain.
Moncalian, G., Cardenes, N., Deribe, Y.L., Spinola-Amilibia, M., Dikic, I., Bravo, J.(2006) J Biological Chem 281: 38845
- PubMed: 17020880
- DOI: https://doi.org/10.1074/jbc.M606411200
- Primary Citation of Related Structures:
2J6F, 2J6K, 2J6O, 2J7I - PubMed Abstract:
The CIN85/CMS (human homologs of mouse SH3KBP1/CD2AP) family of endocytic adaptor proteins has the ability to engage multiple effectors and couple cargo trafficking with the cytoskeleton. CIN85 and CMS (Cas ligand with multiple Src homology 3 (SH3) domains) facilitate the formation of large multiprotein complexes required for an efficient internalization of cell surface receptors. It has recently been shown that c-Cbl/Cbl-b could mediate the formation of a ternary complex between one c-Cbl/Cbl-b molecule and two SH3 domains of CIN85, important for the ability of Cbl to promote epidermal growth factor receptor down-regulation. To further investigate whether multimerization is conserved within the family of adaptor proteins, we have solved the crystal structures of the CMS N-terminal SH3 domain-forming complexes with Cbl-b- and CD2-derived peptides. Together with biochemical evidence, the structures support the notion that, despite clear differences in the interaction surface, both Cbl-b and CD2 can mediate multimerization of N-terminal CMS SH3 domains. Detailed analyses on the interacting surfaces also provide the basis for a differential Cbl-b molecular recognition of CMS and CIN85.
- Signal Transduction Group, Structural Biology and Biocomputing Programme, Centro Nacional de Investigaciones Oncológicas, Melchor Fernández Almagro 3, E-28029 Madrid, Spain.
Organizational Affiliation: