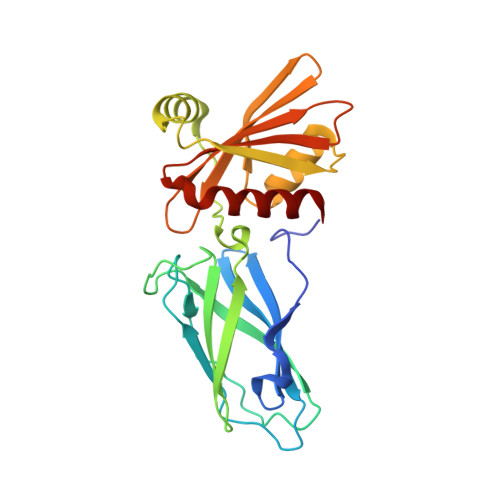
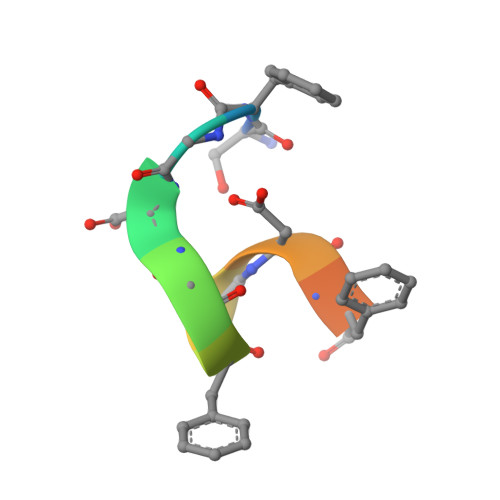
Role of the Ap2 Beta-Appendage Hub in Recruiting Partners for Clathrin-Coated Vesicle Assembly
Schmid, E.M., Ford, M.G.J., Burtey, A., Praefcke, G.J.K., Peak-Chew, S.-Y., Mills, I.G., Benmerah, A., Mcmahon, H.T.(2006) PLoS Biol 4: 262
- PubMed: 16903783
- DOI: https://doi.org/10.1371/journal.pbio.0040262
- Primary Citation of Related Structures:
2IV8, 2IV9 - PubMed Abstract:
Adaptor protein complex 2 alpha and beta-appendage domains act as hubs for the assembly of accessory protein networks involved in clathrin-coated vesicle formation. We identify a large repertoire of beta-appendage interactors by mass spectrometry. These interact with two distinct ligand interaction sites on the beta-appendage (the "top" and "side" sites) that bind motifs distinct from those previously identified on the alpha-appendage. We solved the structure of the beta-appendage with a peptide from the accessory protein Eps15 bound to the side site and with a peptide from the accessory cargo adaptor beta-arrestin bound to the top site. We show that accessory proteins can bind simultaneously to multiple appendages, allowing these to cooperate in enhancing ligand avidities that appear to be irreversible in vitro. We now propose that clathrin, which interacts with the beta-appendage, achieves ligand displacement in vivo by self-polymerisation as the coated pit matures. This changes the interaction environment from liquid-phase, affinity-driven interactions, to interactions driven by solid-phase stability ("matricity"). Accessory proteins that interact solely with the appendages are thereby displaced to areas of the coated pit where clathrin has not yet polymerised. However, proteins such as beta-arrestin (non-visual arrestin) and autosomal recessive hypercholesterolemia protein, which have direct clathrin interactions, will remain in the coated pits with their interacting receptors.
- Medical Research Council Laboratory of Molecular Biology, Cambridge, United Kingdom.
Organizational Affiliation: