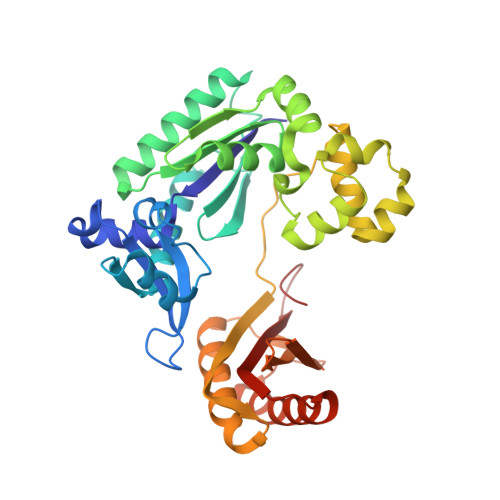
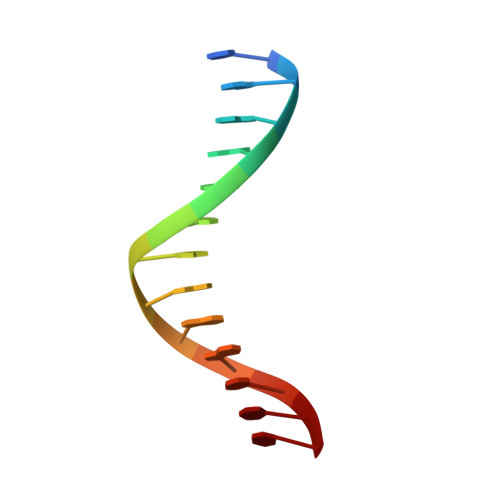
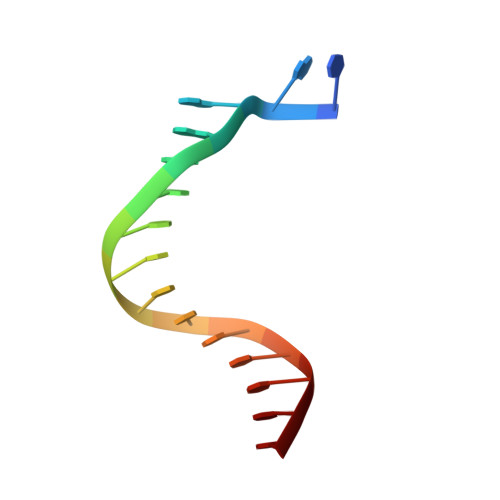
Mechanism of Template-independent Nucleotide Incorporation Catalyzed by a Template-dependent DNA Polymerase.
Fiala, K.A., Brown, J.A., Ling, H., Kshetry, A.K., Zhang, J., Taylor, J.S., Yang, W., Suo, Z.(2007) J Mol Biology 365: 590-602
- PubMed: 17095011
- DOI: https://doi.org/10.1016/j.jmb.2006.10.008
- Primary Citation of Related Structures:
2IMW - PubMed Abstract:
Numerous template-dependent DNA polymerases are capable of catalyzing template-independent nucleotide additions onto blunt-end DNA. Such non-canonical activity has been hypothesized to increase the genomic hypermutability of retroviruses including human immunodeficiency viruses. Here, we employed pre-steady state kinetics and X-ray crystallography to establish a mechanism for blunt-end additions catalyzed by Sulfolobus solfataricus Dpo4. Our kinetic studies indicated that the first blunt-end dATP incorporation was 80-fold more efficient than the second, and among natural deoxynucleotides, dATP was the preferred substrate due to its stronger intrahelical base-stacking ability. Such base-stacking contributions are supported by the 41-fold higher ground-state binding affinity of a nucleotide analog, pyrene nucleoside 5'-triphosphate, which lacks hydrogen bonding ability but possesses four conjugated aromatic rings. A 2.05 A resolution structure of Dpo4*(blunt-end DNA)*ddATP revealed that the base and sugar of the incoming ddATP, respectively, stack against the 5'-base of the opposite strand and the 3'-base of the elongating strand. This unprecedented base-stacking pattern can be applied to subsequent blunt-end additions only if all incorporated dAMPs are extrahelical, leading to predominantly single non-templated dATP incorporation.
- Department of Biochemistry, The Ohio State University, Columbus, OH 43210, USA.
Organizational Affiliation: