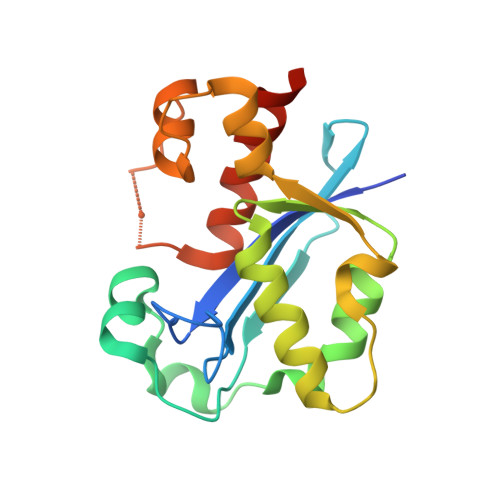
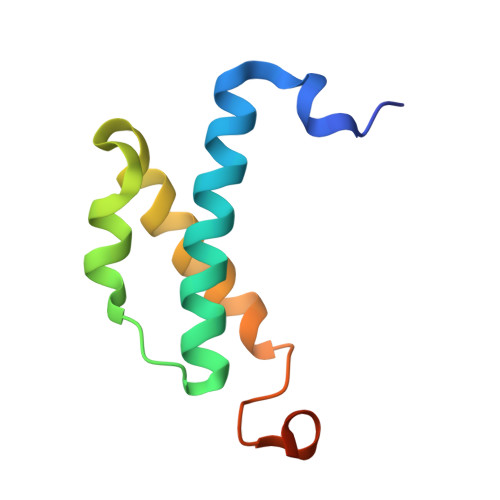
Structure of the Escherichia coli DNA polymerase III epsilon-HOT proofreading complex.
Kirby, T.W., Harvey, S., DeRose, E.F., Chalov, S., Chikova, A.K., Perrino, F.W., Schaaper, R.M., London, R.E., Pedersen, L.C.(2006) J Biological Chem 281: 38466-38471
- PubMed: 16973612
- DOI: https://doi.org/10.1074/jbc.M606917200
- Primary Citation of Related Structures:
2IDO - PubMed Abstract:
The epsilon subunit of Escherichia coli DNA polymerase III possesses 3'-exonucleolytic proofreading activity. Within the Pol III core, epsilon is tightly bound between the alpha subunit (DNA polymerase) and subunit. Here, we present the crystal structure of epsilon in complex with HOT, the bacteriophage P1-encoded homolog of , at 2.1 A resolution. The epsilon-HOT interface is defined by two areas of contact: an interaction of the previously unstructured N terminus of HOT with an edge of the epsilon central beta-sheet as well as interactions between HOT and the catalytically important helix alpha1-loop-helix alpha2 motif of epsilon. This structure provides insight into how HOT and, by implication, may stabilize the epsilon subunit, thus promoting efficient proofreading during chromosomal replication.
- Laboratory of Structural Biology, National Institute of Environmental Health Sciences/NIH, 111 TW Alexander Drive, Research Triangle Park, NC 27709, USA.
Organizational Affiliation: