Structures of a Human Papillomavirus (HPV) E6 Polypeptide Bound to MAGUK Proteins: Mechanisms of Targeting Tumor Suppressors by a High-Risk HPV Oncoprotein.
Zhang, Y., Dasgupta, J., Ma, R.Z., Banks, L., Thomas, M., Chen, X.S.(2007) J Virol 81: 3618-3626
- PubMed: 17267502
- DOI: https://doi.org/10.1128/JVI.02044-06
- Primary Citation of Related Structures:
2I04, 2I0I, 2I0L - PubMed Abstract:
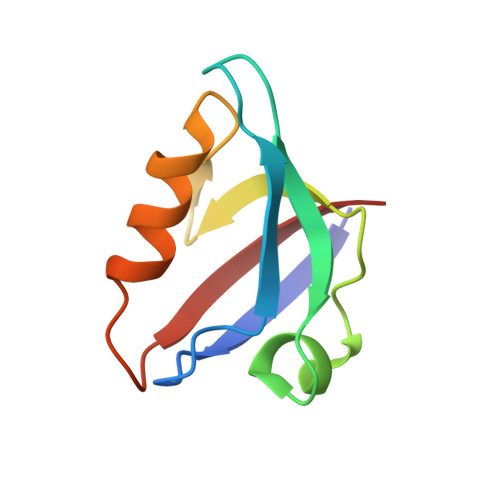
Human papillomavirus (HPV) E6 oncoprotein targets certain tumor suppressors such as MAGI-1 and SAP97/hDlg for degradation. A short peptide at the C terminus of E6 interacts specifically with the PDZ domains of these tumor suppressors, which is a property unique to high-risk HPVs that are associated with cervical cancer. The detailed recognition mechanisms between HPV E6 and PDZ proteins are unclear. To understand the specific binding of cellular PDZ substrates by HPV E6, we have solved the crystal structures of the complexes containing a peptide from HPV18 E6 bound to three PDZ domains from MAGI-1 and SAP97/Dlg. The complex crystal structures reveal novel features of PDZ peptide recognition that explain why high-risk HPV E6 can specifically target these cellular tumor suppressors for destruction. Moreover, a new peptide-binding loop on these PDZs is identified as interacting with the E6 peptide. Furthermore, we have identified an arginine residue, unique to high-risk HPV E6 but outside the canonical core PDZ recognition motif, that plays an important role in the binding of the PDZs of both MAGI-I and SAP97/Dlg, the mutation of which abolishes E6's ability to degrade the two proteins. Finally, we have identified a dimer form of MAGI-1 PDZ domain 1 in the cocrystal structure with E6 peptide, which may have functional relevance for MAGI-1 activity. In addition to its novel insights into the biochemistry of PDZ interactions, this study is important for understanding HPV-induced oncogenesis; this could provide a basis for developing antiviral and anticancer compounds.
- Molecular and Computational Biology, University of Southern California, 1050 Childs Way, MCB201, Los Angeles, CA 90089, USA.
Organizational Affiliation: