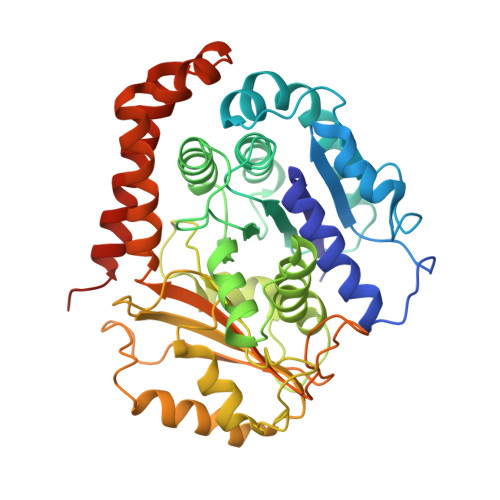
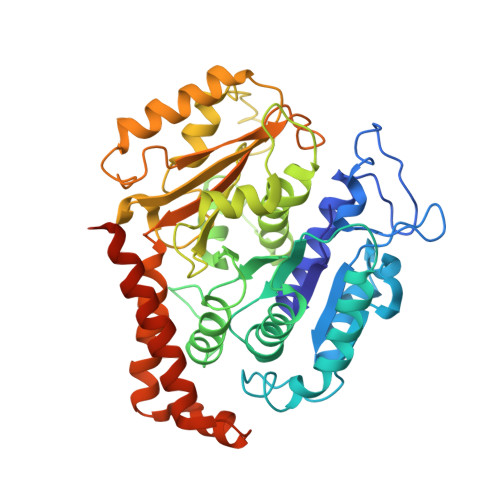
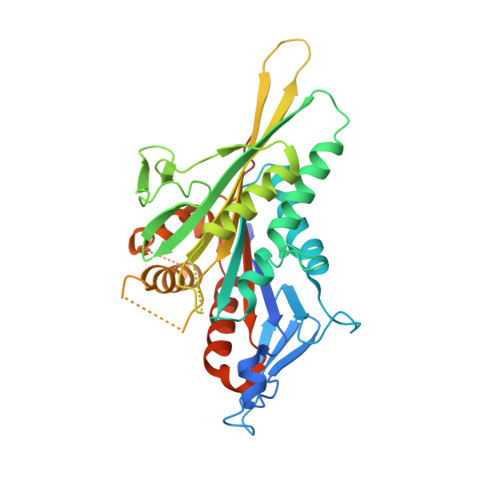
High-resolution cryo-EM maps show the nucleotide binding pocket of KIF1A in open and closed conformations
Kikkawa, M., Hirokawa, N.(2006) EMBO J 25: 4187-4194
- PubMed: 16946706
- DOI: https://doi.org/10.1038/sj.emboj.7601299
- Primary Citation of Related Structures:
2HXF, 2HXH - PubMed Abstract:
Kinesin is an ATP-driven microtubule (MT)-based motor fundamental to organelle transport. Although a number of kinesin crystal structures have been solved, the structural evidence for coupling between the bound nucleotide and the conformation of kinesin is elusive. In addition, the structural basis of the MT-induced ATPase activity of kinesin is not clear because of the absence of the MT in the structure. Here, we report cryo-electron microscopy structures of the monomeric kinesin KIF1A-MT complex in two nucleotide states at about 10 A resolution, sufficient to reveal the secondary structure. These high-resolution maps visualized clear structural changes that suggest a mechanical pathway from the nucleotide to the neck linker via the motor core rotation. In addition, new nucleotide binding pocket conformations are observed that are different from X-ray crystallographic structures; it is closed in the 5'-adenylyl-imidodiphosphate state, but open in the ADP state. These results suggest a structural model of biased diffusion movement of monomeric kinesin motor.
- Department of Cell Biology, Southwestern Medical Center, University of Texas, Dallas, TX, USA.
Organizational Affiliation: