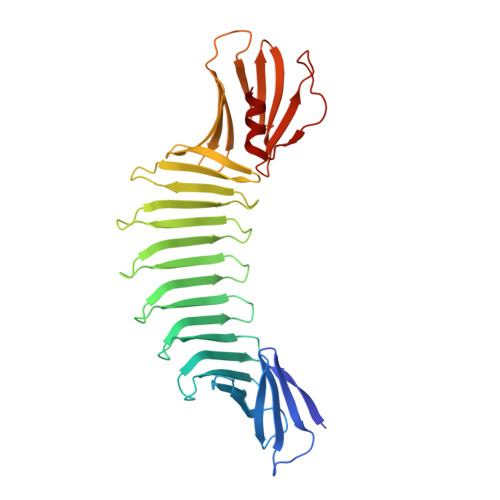
Atomic structures of peptide self-assembly mimics.
Makabe, K., McElheny, D., Tereshko, V., Hilyard, A., Gawlak, G., Yan, S., Koide, A., Koide, S.(2006) Proc Natl Acad Sci U S A 103: 17753-17758
- PubMed: 17093048
- DOI: https://doi.org/10.1073/pnas.0606690103
- Primary Citation of Related Structures:
2AF5, 2FKG, 2FKJ, 2HKD - PubMed Abstract:
Although the beta-rich self-assemblies are a major structural class for polypeptides and the focus of intense research, little is known about their atomic structures and dynamics due to their insoluble and noncrystalline nature. We developed a protein engineering strategy that captures a self-assembly segment in a water-soluble molecule. A predefined number of self-assembling peptide units are linked, and the beta-sheet ends are capped to prevent aggregation, which yields a mono-dispersed soluble protein. We tested this strategy by using Borrelia outer surface protein (OspA) whose single-layer beta-sheet located between two globular domains consists of two beta-hairpin units and thus can be considered as a prototype of self-assembly. We constructed self-assembly mimics of different sizes and determined their atomic structures using x-ray crystallography and NMR spectroscopy. Highly regular beta-sheet geometries were maintained in these structures, and peptide units had a nearly identical conformation, supporting the concept that a peptide in the regular beta-geometry is primed for self-assembly. However, we found small but significant differences in the relative orientation between adjacent peptide units in terms of beta-sheet twist and bend, suggesting their inherent flexibility. Modeling shows how this conformational diversity, when propagated over a large number of peptide units, can lead to a substantial degree of nanoscale polymorphism of self-assemblies.
- Department of Biochemistry and Molecular Biology, University of Chicago, Chicago, IL 60637, USA.
Organizational Affiliation: