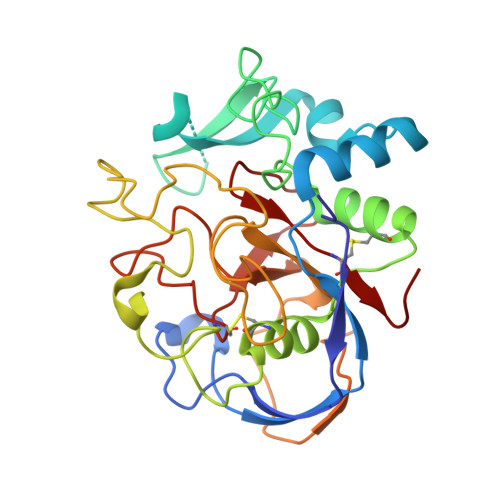
Probing the oxygen-binding site of the human formylglycine-generating enzyme using halide ions.
Roeser, D., Schmidt, B., Preusser-Kunze, A., Rudolph, M.G.(2007) Acta Crystallogr D Biol Crystallogr 63: 621-627
- PubMed: 17452787
- DOI: https://doi.org/10.1107/S0907444907009961
- Primary Citation of Related Structures:
2HI8, 2HIB - PubMed Abstract:
The catalytic residue in sulfatases is a unique formylglycine that is post-translationally generated by oxidation of a cysteine or serine precursor. Molecular oxygen oxidizes the cysteine precursor in eukaryotic sulfatases, a reaction that is catalysed by the formylglycine-generating enzyme FGE. Previously, FGE was crystallized in complex with a chloride ion which, based on its similar polarizability and hydrophobicity, indicates the site of molecular oxygen binding. Here, two structures of FGE in complex with bromide and iodide were determined in order to further delineate the volume and stereochemical restraints of the oxygen-binding site for potential reaction intermediates. Anomalous difference density maps unambiguously assigned the nature of the halide ions. Unexpectedly, data collected at a wavelength of 1.54 A from the iodide-containing crystal and data collected at a wavelength of 0.8 A from a bromide-containing crystal were sufficient for SIRAS phasing.
- Department of Molecular Structural Biology, University of Göttingen, D-37077 Göttingen, Germany.
Organizational Affiliation: