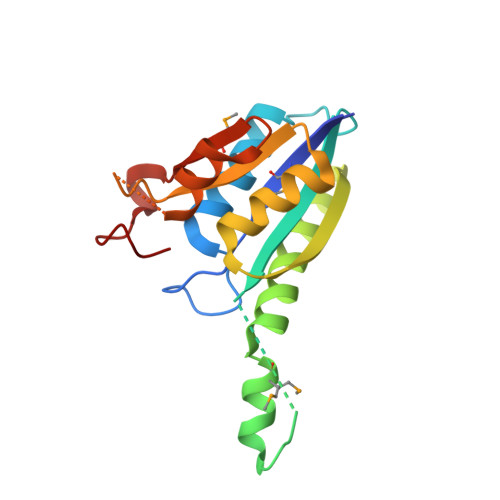
Crystal structure of the protein At3g01520, a eukaryotic universal stress protein-like protein from arabidopsis thaliana in complex with AMP.
Kim, D.O.J., Bitto, E., Bingman, C.A., Kim, H.J., Han, B.W., Phillips, G.N.(2015) Proteins 83: 1368-1373
- PubMed: 25921306
- DOI: https://doi.org/10.1002/prot.24821
- Primary Citation of Related Structures:
2GM3 - PubMed Abstract:
Members of the universal stress protein (USP) family are conserved in a phylogenetically diverse range of prokaryotes, fungi, protists, and plants and confer abilities to respond to a wide range of environmental stresses. Arabidopsis thaliana contains 44 USP domain-containing proteins, and USP domain is found either in a small protein with unknown physiological function or in an N-terminal portion of a multi-domain protein, usually a protein kinase. Here, we report the first crystal structure of a eukaryotic USP-like protein encoded from the gene At3g01520. The crystal structure of the protein At3g01520 was determined by the single-wavelength anomalous dispersion method and refined to an R factor of 21.8% (Rfree = 26.1%) at 2.5 Å resolution. The crystal structure includes three At3g01520 protein dimers with one AMP molecule bound to each protomer, comprising a Rossmann-like α/β overall fold. The bound AMP and conservation of residues in the ATP-binding loop suggest that the protein At3g01520 also belongs to the ATP-binding USP subfamily members.
- Research Institute of Pharmaceutical Sciences, College of Pharmacy, Seoul National University, Seoul, 151-742, Korea.
Organizational Affiliation: