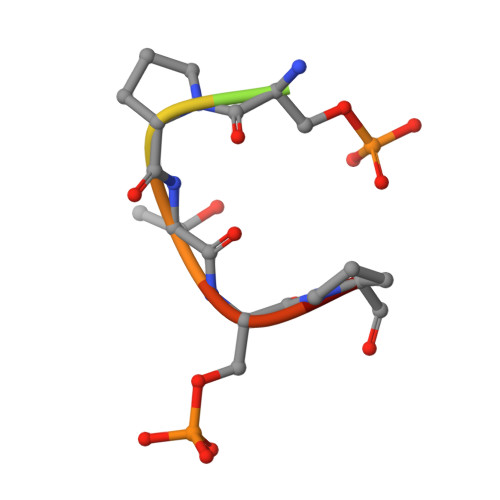
Determinants for dephosphorylation of the RNA polymerase II C-terminal domain by Scp1.
Zhang, Y., Kim, Y., Genoud, N., Gao, J., Kelly, J.W., Pfaff, S.L., Gill, G.N., Dixon, J.E., Noel, J.P.(2006) Mol Cell 24: 759-770
- PubMed: 17157258
- DOI: https://doi.org/10.1016/j.molcel.2006.10.027
- Primary Citation of Related Structures:
2GHQ, 2GHT - PubMed Abstract:
Phosphorylation and dephosphorylation of the C-terminal domain (CTD) of RNA polymerase II (Pol II) represent a critical regulatory checkpoint for transcription. Transcription initiation requires Fcp1/Scp1-mediated dephosphorylation of phospho-CTD. Fcp1 and Scp1 belong to a family of Mg2+ -dependent phosphoserine (P.Ser)/phosphothreonine (P.Thr)-specific phosphatases. We recently showed that Scp1 is an evolutionarily conserved regulator of neuronal gene silencing. Here, we present the X-ray crystal structures of a dominant-negative form of human Scp1 (D96N mutant) bound to mono- and diphosphorylated peptides encompassing the CTD heptad repeat (Y1S2P3T4S5P6S7). Moreover, kinetic and thermodynamic analyses of Scp1-phospho-CTD peptide complexes support the structures determined. This combined structure-function analysis discloses the residues in Scp1 involved in CTD binding and its preferential dephosphorylation of P.Ser5 of the CTD heptad repeat. Moreover, these results provide a template for the design of specific inhibitors of Scp1 for the study of neuronal stem cell development.
- Howard Hughes Medical Institute, Jack H. Skirball Center for Chemical Biology and Proteomics, The Salk Institute for Biological Studies, La Jolla, California 92037.
Organizational Affiliation: