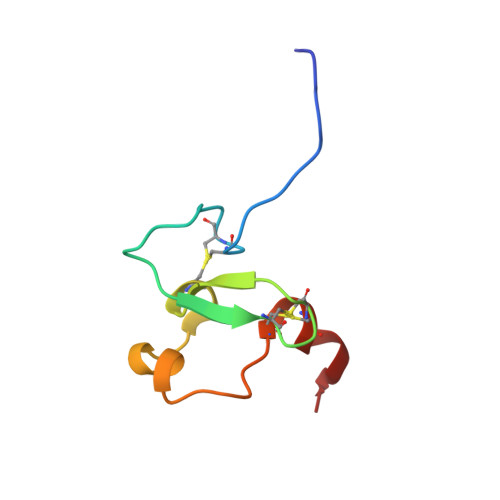
Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF.
Li, H., Ilin, S., Wang, W., Duncan, E.M., Wysocka, J., Allis, C.D., Patel, D.J.(2006) Nature 442: 91-95
- PubMed: 16728978
- DOI: https://doi.org/10.1038/nature04802
- Primary Citation of Related Structures:
2F6J, 2F6N, 2FSA, 2FUI, 2FUU - PubMed Abstract:
Mono-, di- and trimethylated states of particular histone lysine residues are selectively found in different regions of chromatin, thereby implying specialized biological functions for these marks ranging from heterochromatin formation to X-chromosome inactivation and transcriptional regulation. A major challenge in chromatin biology has centred on efforts to define the connection between specific methylation states and distinct biological read-outs impacting on function. For example, histone H3 trimethylated at lysine 4 (H3K4me3) is associated with transcription start sites of active genes, but the molecular 'effectors' involved in specific recognition of H3K4me3 tails remain poorly understood. Here we demonstrate the molecular basis for specific recognition of H3(1-15)K4me3 (residues 1-15 of histone H3 trimethylated at K4) by a plant homeodomain (PHD) finger of human BPTF (bromodomain and PHD domain transcription factor), the largest subunit of the ATP-dependent chromatin-remodelling complex, NURF (nucleosome remodelling factor). We report on crystallographic and NMR structures of the bromodomain-proximal PHD finger of BPTF in free and H3(1-15)K4me3-bound states. H3(1-15)K4me3 interacts through anti-parallel beta-sheet formation on the surface of the PHD finger, with the long side chains of arginine 2 (R2) and K4me3 fitting snugly in adjacent pre-formed surface pockets, and bracketing an invariant tryptophan. The observed stapling role by non-adjacent R2 and K4me3 provides a molecular explanation for H3K4me3 site specificity. Binding studies establish that the BPTF PHD finger exhibits a modest preference for K4me3- over K4me2-containing H3 peptides, and discriminates against monomethylated and unmodified counterparts. Furthermore, we identified key specificity-determining residues from binding studies of H3(1-15)K4me3 with PHD finger point mutants. Our findings call attention to the PHD finger as a previously uncharacterized chromatin-binding module found in a large number of chromatin-associated proteins.
- Structural Biology Program, Memorial Sloan-Kettering Cancer Center, New York, New York 10021, USA.
Organizational Affiliation: