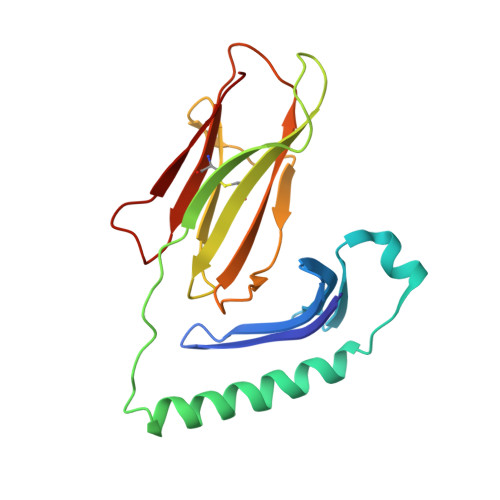
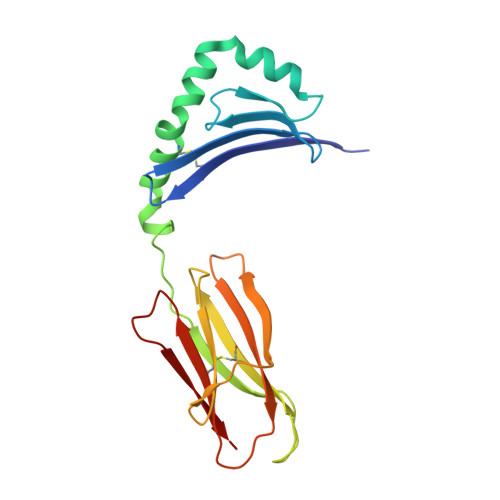
Crystallographic Structure of a Rheumatoid Arthritis MHC Susceptibility Allele, HLA-DR1 (DRB1*0101), Complexed with the Immunodominant Determinant of Human Type II Collagen.
Rosloniec, E.F., Ivey, R.A., Whittington, K.B., Kang, A.H., Park, H.W.(2006) J Immunol 177: 3884-3892
- PubMed: 16951351
- DOI: https://doi.org/10.4049/jimmunol.177.6.3884
- Primary Citation of Related Structures:
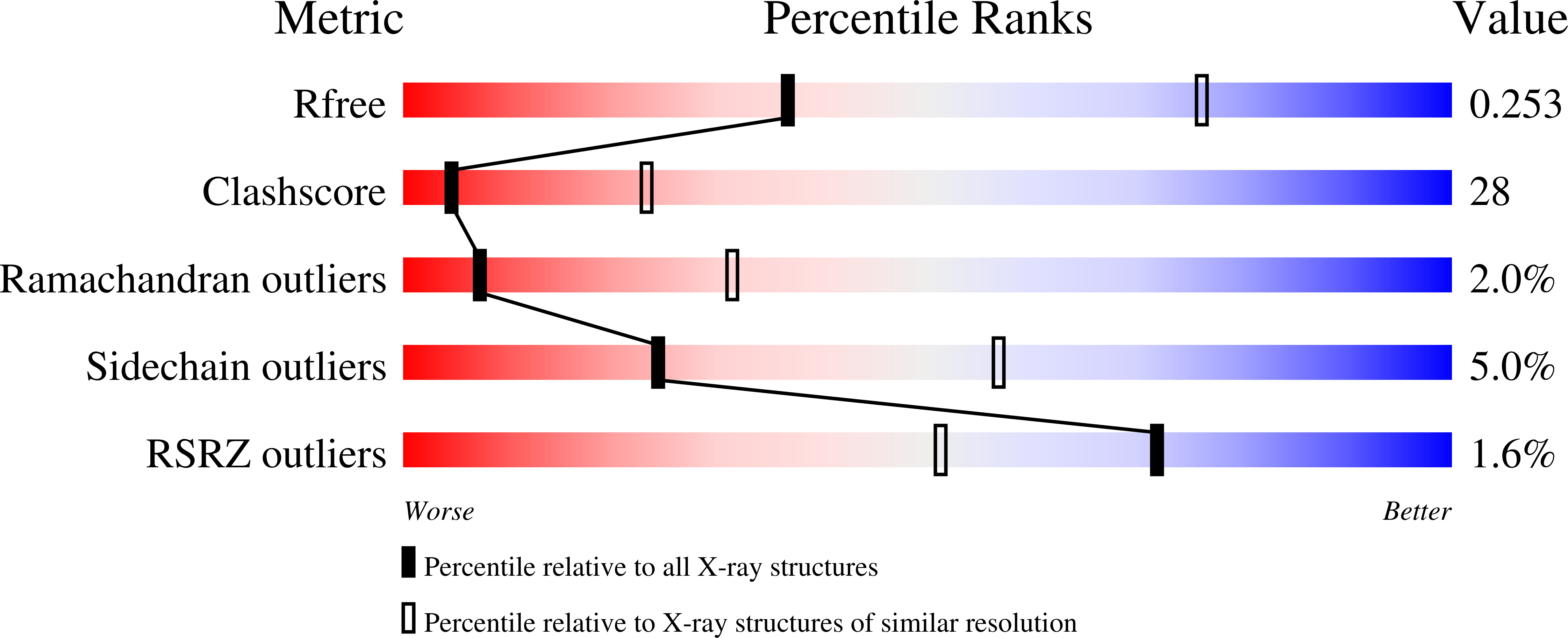
2FSE - PubMed Abstract:
The expression of HLA-DR1 (DRB1*0101) is associated with an enhanced risk for developing rheumatoid arthritis (RA). To study its function, we have solved the three-dimensional structure of HLA-DR1 complexed with a candidate RA autoantigen, the human type II collagen peptide CII (259-273). Based on these structural data, the CII peptide is anchored by Phe263 at the P1 position and Glu266 at P4. Surprisingly, the Lys at the P2 position appears to play a dual role by participating in peptide binding via interactions with DRB1-His81 and Asn82, and TCR interaction, based on functional assays. The CII peptide is also anchored by the P4 Glu266 residue through an ionic interaction with DRB1-Arg71 and Glu28. Participation of DRB1-Arg71 is significant because it is part of the shared epitope expressed by DR alleles associated with RA susceptibility. Potential anchor residues at P6 and P9 of the CII peptide are both Gly, and the lack of side chains at these positions appears to result in both a narrower binding groove with the peptide protruding out of the groove at this end of the DR1 molecule. From the TCR perspective, the P2-Lys264, P5-Arg267, and P8-Lys270 residues are all oriented away from the binding groove and collectively represent a positive charged interface for CII-specific TCR binding. Comparison of the DR1-CII structure to a DR1-hemagglutinin peptide structure revealed that the binding of these two peptides generates significantly different interfaces for the interaction with their respective Ag-specific TCRs.
- Veterans Affairs Medical Center, Memphis, TN 38104, USA.
Organizational Affiliation: