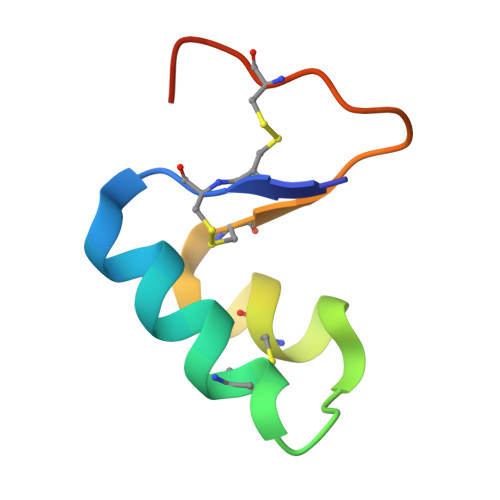
Role of a salt bridge in the model protein crambin explored by chemical protein synthesis: X-ray structure of a unique protein analogue, [V15A]crambin-alpha-carboxamide.
Bang, D., Tereshko, V., Kossiakoff, A.A., Kent, S.B.(2009) Mol Biosyst 5: 750-756
- PubMed: 19562114
- DOI: https://doi.org/10.1039/b903610e
- Primary Citation of Related Structures:
2FD7, 2FD9 - PubMed Abstract:
We have used total chemical synthesis to prepare [V15A]crambin-alpha-carboxamide, a unique protein analogue that eliminates a salt bridge between the delta-guanidinium of the Arg(10) side chain and the alpha-carboxylate of Asn(46) at the C-terminus of the polypeptide chain. This salt bridge is thought to be important for the folding and stability of the crambin protein molecule. Folding, with concomitant disulfide bond formation, of the fully reduced [V15A]crambin-alpha-carboxamide polypeptide was less efficient than folding/disulfide formation for the [V15A]crambin polypeptide under a standard set of conditions. To probe the origin of this less efficient folding/disulfide bond formation, we separately crystallized purified synthetic [V15A]crambin-alpha-carboxamide and chemically synthesized [V15A]crambin and solved their X-ray structures. The crystal structure of [V15A]crambin-alpha-carboxamide showed that elimination of the Arg(10)-Asn(46) salt bridge caused disorder of the C-terminal region of the polypeptide chain and affected the overall 'tightness' of the structure of the protein molecule. These studies, enabled by chemical protein synthesis, strongly suggest that in native crambin the Arg(10)-Asn(46) salt bridge contributes to efficient formation of correct disulfide bonds and also to the well-ordered structure of the protein molecule.
- Institute for Biophysical Dynamics, The University of Chicago, Chicago, IL 60637, USA.
Organizational Affiliation: