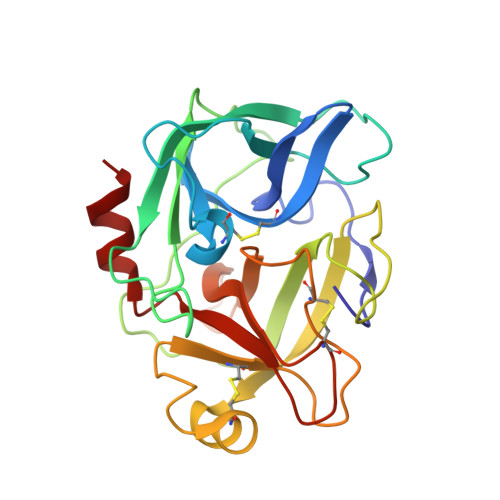
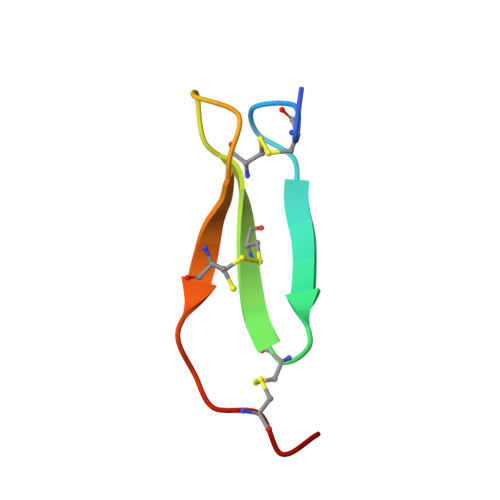
Enzyme:Substrate Hydrogen Bond Shortening during the Acylation Phase of Serine Protease Catalysis.
Fodor, K., Harmat, V., Neutze, R., Szilagyi, L., Graf, L., Katona, G.(2006) Biochemistry 45: 2114-2121
- PubMed: 16475800
- DOI: https://doi.org/10.1021/bi0517133
- Primary Citation of Related Structures:
2F91 - PubMed Abstract:
Atomic resolution (
- Biotechnology Research Group of the Hungarian Academy of Sciences, Pázmány Street 1/C, 1117 Budapest, Hungary.
Organizational Affiliation: