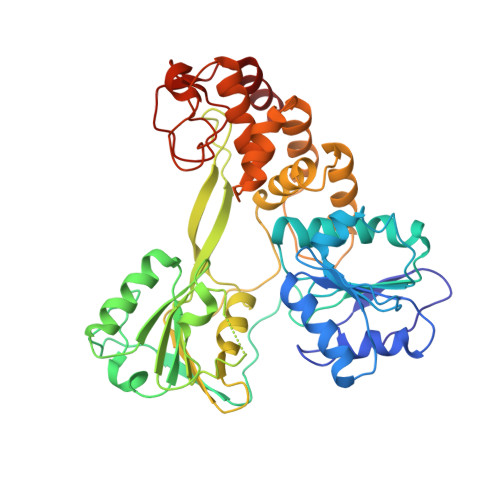
Structural and biological identification of residues on the surface of NS3 helicase required for optimal replication of the hepatitis C virus
Mackintosh, S.G., Lu, J.Z., Jordan, J.B., Harrison, M.K., Sikora, B., Sharma, S.D., Cameron, C.E., Raney, K.D., Sakon, J.(2006) J Biological Chem 281: 3528-3535
- PubMed: 16306038
- DOI: https://doi.org/10.1074/jbc.M512100200
- Primary Citation of Related Structures:
2F55 - PubMed Abstract:
The hepatitis C virus (HCV) nonstructural protein 3 (NS3) is a multifunctional enzyme with serine protease and DEXH/D-box helicase domains. A crystal structure of the NS3 helicase domain (NS3h) was generated in the presence of a single-stranded oligonucleotide long enough to accommodate binding of two molecules of enzyme. Several amino acid residues at the interface of the two NS3h molecules were identified that appear to mediate a protein-protein interaction between domains 2 and 3 of adjacent molecules. Mutations were introduced into domain 3 to disrupt the putative interface and subsequently examined using an HCV subgenomic replicon, resulting in significant reduction in replication capacity. The mutations in domain 3 were then examined using recombinant NS3h in biochemical assays. The mutant enzyme showed RNA binding and RNA-stimulated ATPase activity that mirrored wild type NS3h. In DNA unwinding assays under single turnover conditions, the mutant NS3h exhibited a similar unwinding rate and only approximately 2-fold lower processivity than wild type NS3h. Overall biochemical activities of the mutant NS3h were similar to the wild type enzyme, which was not reflective of the large reduction in HCV replicative capacity observed in the biological experiment. Hence, the biological results suggest that the known biochemical properties associated with the helicase activity of NS3h do not reveal all of the likely biological roles of NS3 during HCV replication. Domain 3 of NS3 is implicated in protein-protein interactions that are necessary for HCV replication.
- Department of Biochemistry and Molecular Biology, University of Arkansas for Medical Sciences, Little Rock, Arkansas 72205, USA.
Organizational Affiliation: