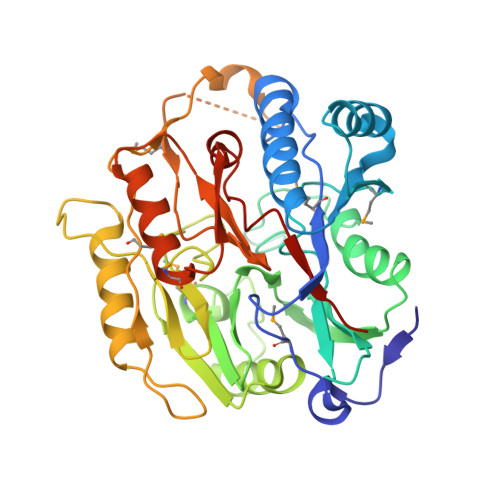
X-ray structure of putative agmatine deiminase Q8DW17, Northeast Structural Genomics target SmR6.
Kuzin, A.P., Chen, Y., Vorobiev, S.M., Su, M., Forouhar, F., Acton, T., Xiao, R., Conover, K., Ma, L.-C., Kellie, R., Cunningham, K.E., Montelione, G.T., Hunt, J.F., Tong, L.To be published.