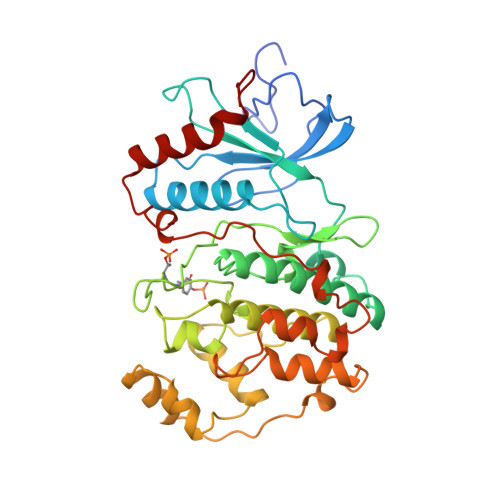
Activation mechanism of the MAP kinase ERK2 by dual phosphorylation.
Canagarajah, B.J., Khokhlatchev, A., Cobb, M.H., Goldsmith, E.J.(1997) Cell 90: 859-869
- PubMed: 9298898
- DOI: https://doi.org/10.1016/s0092-8674(00)80351-7
- Primary Citation of Related Structures:
2ERK - PubMed Abstract:
The structure of the active form of the MAP kinase ERK2 has been solved, phosphorylated on a threonine and a tyrosine residue within the phosphorylation lip. The lip is refolded, bringing the phosphothreonine and phosphotyrosine into alignment with surface arginine-rich binding sites. Conformational changes occur in the lip and neighboring structures, including the P+1 site, the MAP kinase insertion, the C-terminal extension, and helix C. Domain rotation and remodeling of the proline-directed P+1 specificity pocket account for the activation. The conformation of the P+1 pocket is similar to a second proline-directed kinase, CDK2-CyclinA, thus permitting the origin of this specificity to be defined. Conformational changes outside the lip provide loci at which the state of phosphorylation can be felt by other cellular components.
- Department of Biochemistry, The University of Texas Southwestern Medical Center at Dallas, 75235-9050, USA.
Organizational Affiliation: