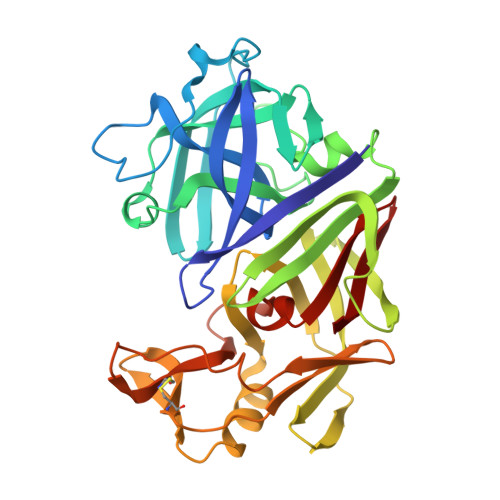
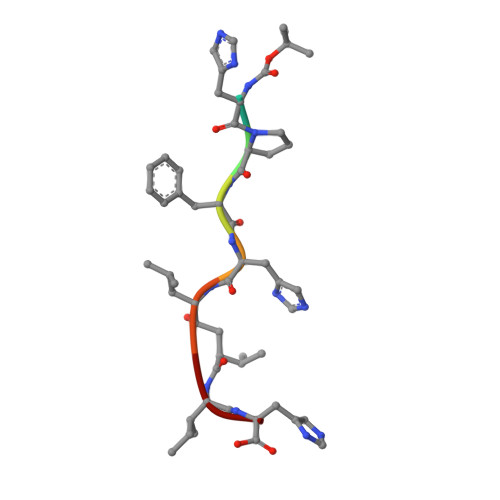
X-ray analyses of aspartic proteinases. III Three-dimensional structure of endothiapepsin complexed with a transition-state isostere inhibitor of renin at 1.6 A resolution.
Veerapandian, B., Cooper, J.B., Sali, A., Blundell, T.L.(1990) J Mol Biology 216: 1017-1029
- PubMed: 2266553
- DOI: https://doi.org/10.1016/S0022-2836(99)80017-5
- Primary Citation of Related Structures:
2ER7 - PubMed Abstract:
The aspartic proteinase, endothiapepsin (EC 3.4.23.6), was complexed with a highly potent renin inhibitor, H-261 (t-Boc-His-Pro-Phe-His-LeuOHVal-Ile-His), where OH denotes a hydroxyethylene (-(S) CHOH-CH2-) transition-state isostere in the scissile bond surrogate. Crystals were grown in a form that has the same space group P2(1) as the uncomplexed enzyme, but with a 10 A decrease in the length of the alpha-axis and a 13 degrees decrease in the beta-angle. X-ray data have been collected to a resolution of 1.6 A. The rotation and translation parameters defining the position of the enzyme in the unit cell were determined previously using another enzyme-inhibitor complex that crystallized isomorphously with that of H-261. The molecule was refined using restrained least-squares refinement and the positions of non-hydrogen atoms of the inhibitor and water molecules were defined by difference Fourier techniques. The enzyme-inhibitor complex and 322 water molecules were further refined to a crystallographic R-factor of 0.14. Apart from a small rigid group rotation of a domain comprising residues 190 to 302 and small movements in the flap, there is little difference in conformation between the complexed and uncomplexed forms of the enzyme. The inhibitor is bound in an extended conformation along the active site cleft, and the hydroxyl group of the hydroxyethylene moiety is hydrogen-bonded to both catalytic aspartate carboxylates. The complex is stabilized by hydrogen bonds between the main-chain of the inhibitor and the enzyme. All side-chains of the inhibitor are in van der Waals' contact with groups in the enzyme and define a series of specificity pockets along the active site cleft. The study provides useful clues as to how this potent renin inhibitor (IC50 value of 0.7 x 10(-9) M) may bind renin. In particular it defines the interactions of the hydroxyethylene transition-state isostere with the enzyme more precisely than has been previously possible and therefore provides a useful insight into interactions in the transition state complex.
- Department of Crystallography, Birkbeck College, London, U.K.
Organizational Affiliation: