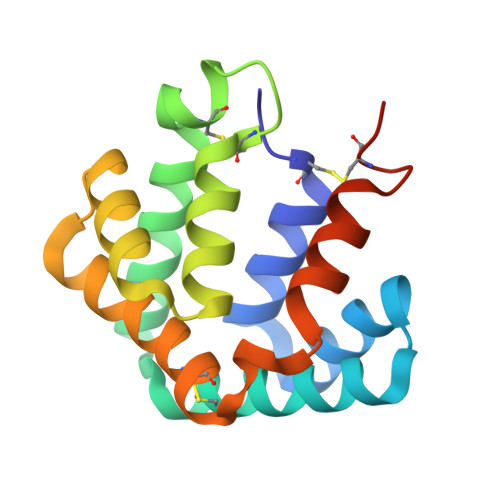
Structural characterization of the tetrameric form of the major cat allergen Fel d 1
Kaiser, L., Velickovic, T.C., Badia-Martinez, D., Adedoyin, J., Thunberg, S., Hallen, D., Berndt, K., Gronlund, H., Gafvelin, G., van Hage, M., Achour, A.(2007) J Mol Biology 370: 714-727
- PubMed: 17543334
- DOI: https://doi.org/10.1016/j.jmb.2007.04.074
- Primary Citation of Related Structures:
2EJN - PubMed Abstract:
Felis domesticus allergen 1(Fel d 1) is a 35 kDa tetrameric glycoprotein formed by two heterodimers which elicits IgE responses in 95% of patients with allergy to cat. We have previously established in vitro conditions for the appropriate folding of recombinant Fel d 1 using a direct linkage of chain 1 to chain 2 (construct Fel d 1 (1+2)) and chain 2 to chain 1 (construct Fel d 1 (2+1)). Although the crystal structure of Fel d 1 (2+1) revealed a striking structural similarity to that of uteroglobin, a steroid-inducible cytokine-like molecule with anti-inflammatory and immunomodulatory properties, no functional tetrameric form of Fel d 1 could be identified. Here we present the crystal structure of the Fel d 1 (1+2) tetramer at 1.6 A resolution. Interestingly, the crystal structure of tetrameric Fel d 1 reveals two different calcium-binding sites. Symmetrically positioned on each side of the Fel d 1 tetramer, the external Ca(2+)-binding sites correspond to a putative Ca(2+)-binding site previously suggested for uteroglobin. The second Ca(2+)-binding site lies within the dimerization interface, stabilizing the formation of the Fel d 1 tetramer, and inducing important local conformational changes that directly govern the shape of two water-filled cavities. The crystal structure suggests a potential portal for an unknown ligand. Alternatively, the two cavities could be used by the allergen as a conditional inner space allowing for the spatial rearrangement of centrally localized side-chains, such as Asp130, without altering the overall fold of the molecule. The striking structural similarity of the major cat allergen to uteroglobin, coupled to the identification in the present study of a common Ca(2+)-binding site, let us speculate that Fel d 1 could provoke an allergic response through the modulation of phospholipase A2, by sequestering Ca ions in a similar manner as previously suggested for uteroglobin.
- Center for Infectious Medicine, Department of Medicine, Karolinska Institutet, Karolinska University Hospital Huddinge, Stockholm, Sweden.
Organizational Affiliation: