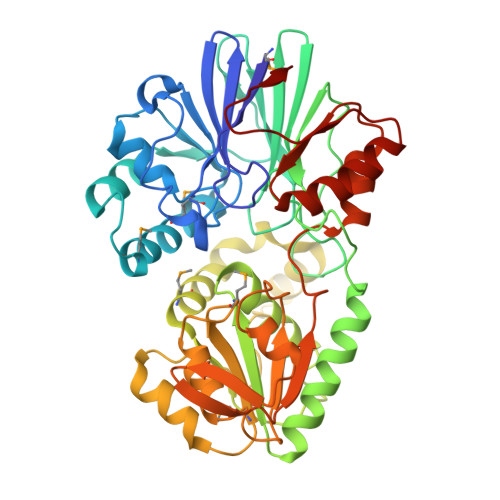
Crystal structure of TTHA0252 from Thermus thermophilus HB8, a RNA degradation protein of the metallo-beta-lactamase superfamily
Ishikawa, H., Nakagawa, N., Kuramitsu, S., Masui, R.(2006) J Biochem 140: 535-542
- PubMed: 16945939
- DOI: https://doi.org/10.1093/jb/mvj183
- Primary Citation of Related Structures:
2DKF - PubMed Abstract:
In bacterial RNA metabolism, mRNA degradation is an important process for gene expression. Recently, a novel ribonuclease (RNase), belonging to the beta-CASP family within the metallo-beta-lactamase superfamily, was identified as a functional homologue of RNase E, a major component for mRNA degradation in Escherichia coli. Here, we have determined the crystal structure of TTHA0252 from Thermus thermophilus HB8, which represents the first report of the tertiary structure of a beta-CASP family protein. TTHA0252 comprises two separate domains: a metallo-beta-lactamase domain and a "clamp" domain. The active site of the enzyme is located in a cleft between the two domains, which includes two zinc ions coordinated by seven conserved residues. Although this configuration is similar to those of other beta-lactamases, TTHA0252 has one conserved His residue characteristic of the beta-CASP family as a ligand. We also detected nuclease activity of TTHA0252 against rRNAs of T. thermophilus. Our results reveal structural and functional aspects of novel RNase E-like enzymes with a beta-CASP fold.
- Department of Biological Sciences, Graduate School of Science, Osaka University, Toyonaka, Osaka 560-0043, Japan.
Organizational Affiliation: