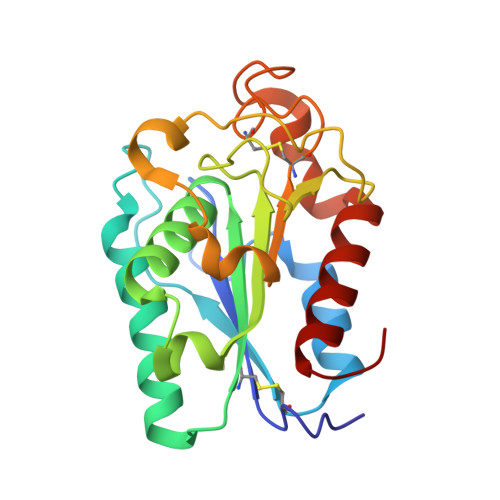
Crystal structure and enhanced activity of a cutinase-like enzyme from Cryptococcus sp. strain S-2
Kodama, Y., Masaki, K., Kondo, H., Suzuki, M., Tsuda, S., Nagura, T., Shimba, N., Suzuki, E., Iefuji, H.(2009) Proteins 77: 710-717
- PubMed: 19544571
- DOI: https://doi.org/10.1002/prot.22484
- Primary Citation of Related Structures:
2CZQ - PubMed Abstract:
The structural and enzymatic characteristics of a cutinase-like enzyme (CLE) from Cryptococcus sp. strain S-2, which exhibits remote homology to a lipolytic enzyme and a cutinase from the fungus Fusarium solani (FS cutinase), were compared to investigate the unique substrate specificity of CLE. The crystal structure of CLE was solved to a 1.05 A resolution. Moreover, hydrolysis assays demonstrated the broad specificity of CLE for short and long-chain substrates, as well as the preferred specificity of FS cutinase for short-chain substrates. In addition, site-directed mutagenesis was performed to increase the hydrolysis activity on long-chain substrates, indicating that the hydrophobic aromatic residues are important for the specificity to the long-chain substrate. These results indicate that hydrophobic residues, especially the aromatic ones exposed to solvent, are important for retaining lipase activity.
- Institute of Life Sciences, Ajinomoto Co., Inc., Kawasaki-ku, Kawasaki-shi 210-8681, Japan.
Organizational Affiliation: