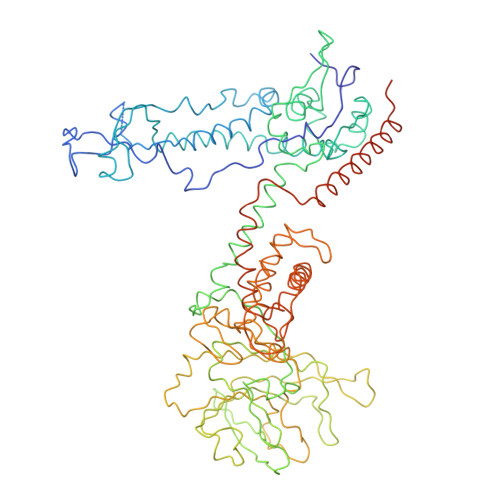
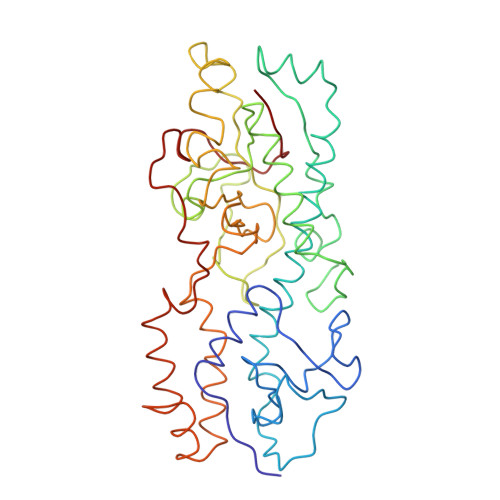
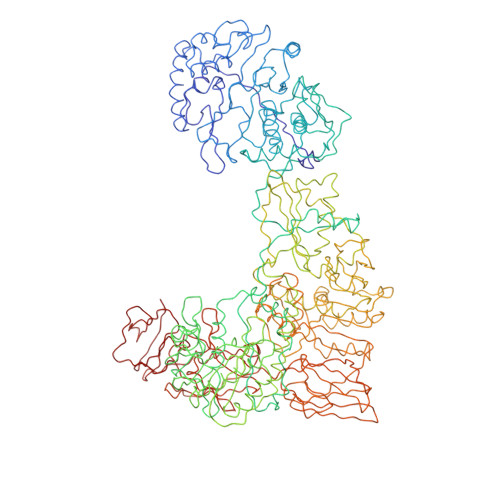
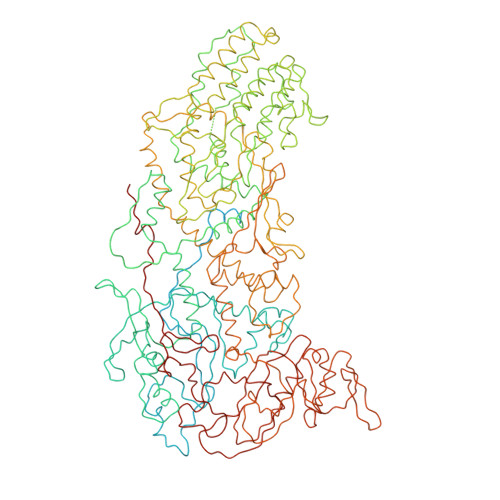
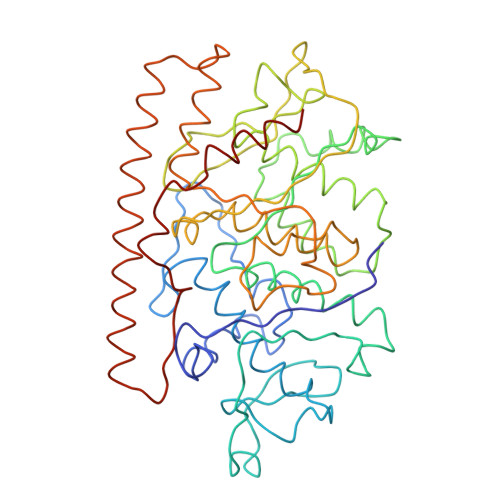
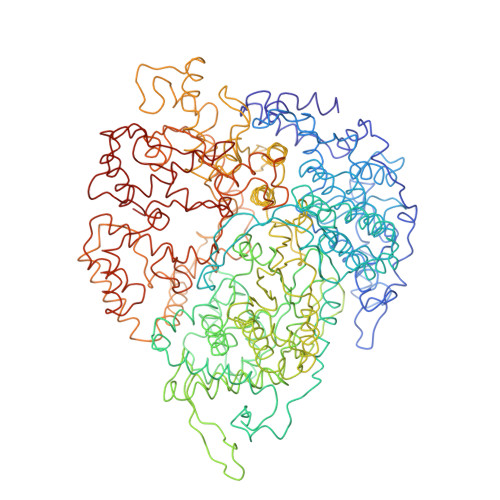
Features of reovirus outer capsid protein mu1 revealed by electron cryomicroscopy and image reconstruction of the virion at 7.0 Angstrom resolution.
Zhang, X., Ji, Y., Zhang, L., Harrison, S.C., Marinescu, D.C., Nibert, M.L., Baker, T.S.(2005) Structure 13: 1545-1557
- PubMed: 16216585
- DOI: https://doi.org/10.1016/j.str.2005.07.012
- Primary Citation of Related Structures:
2CSE - PubMed Abstract:
Reovirus is a useful model for addressing the molecular basis of membrane penetration by one of the larger nonenveloped animal viruses. We now report the structure of the reovirus virion at approximately 7.0 A resolution as obtained by electron cryomicroscopy and three-dimensional image reconstruction. Several features of the myristoylated outer capsid protein mu1, not seen in a previous X-ray crystal structure of the mu1-sigma3 heterohexamer, are evident in the virion. These features appear to be important for stabilizing the outer capsid, regulating the conformational changes in mu1 that accompany perforation of target membranes, and contributing directly to membrane penetration during cell entry.
- Department of Biological Sciences, Purdue University, West Lafayette, Indiana 47907, USA.
Organizational Affiliation: