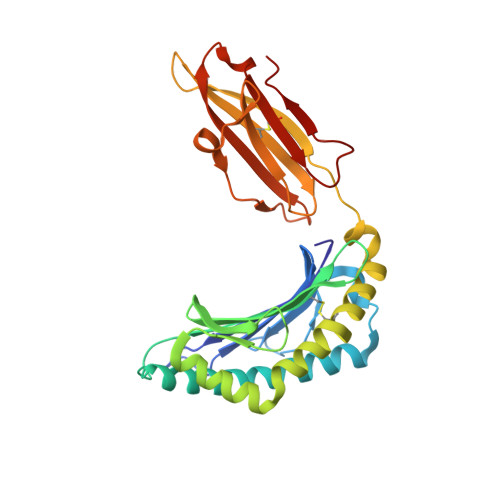
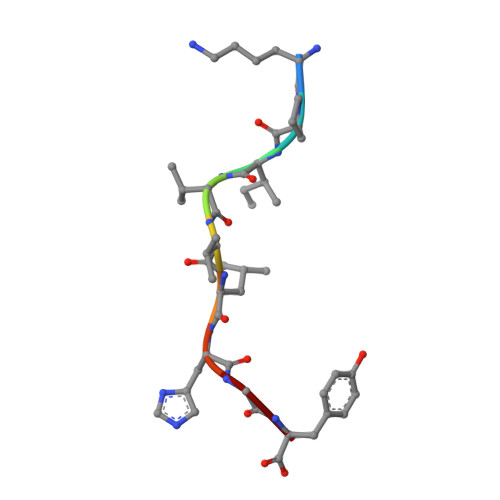
The Structure of the Human Allo-Ligand Hla-B3501 in Complex with a Cytochrome P450 Peptide: Steric Hindrance Influences Tcr Allo-Recognition.
Hourigan, C.S., Harkiolaki, M., Peterson, N.A., Bell, J.I., Jones, E.Y., O'Callaghan, C.A.(2006) Eur J Immunol 36: 3288
- PubMed: 17109469
- DOI: https://doi.org/10.1002/eji.200636234
- Primary Citation of Related Structures:
2CIK - PubMed Abstract:
Virus-specific T cell populations have been implicated in allo-recognition. The subdominant T cell receptor JL12 recognizes both HLA-B*0801 presenting the Epstein-Barr virus-derived peptide FLRGRAYGL and also HLA-B*3501 presenting the cytochrome p450 self peptide KPIVVLHGY. This cross-reactivity could promote the rejection of HLA-B*3501-positive cells in Epstein-Barr virus-exposed HLA-B*0801 recipients. LC13, the dominant TCR against the HLA-B*0801:FLRGRAYGL complex, fails to recognize HLA-B*3501:KPIVVLHGY. We report the 1.75-Angstrom resolution crystal structure of the human allo-ligand HLA-B*3501:KPIVVLHGY. Similarities between this structure and that of HLA-B*0801:FLRGRAYGL may facilitate cross-recognition by JL12. Moreover, the elevated peptide position in HLA-B*3501:KPIVVLHGY would provide steric hindrance to LC13, preventing it from interacting in the manner in which it interacts with HLA-B*0801:FLRGRAYGL. These findings are relevant to understanding the basis of T cell cross-reactivity in allo-recognition, optimal transplant donor-recipient matching and developing specific molecular inhibitors of allo-recognition.
- Nuffield Department of Medicine, John Radcliffe Hospital, Oxford, UK.
Organizational Affiliation: