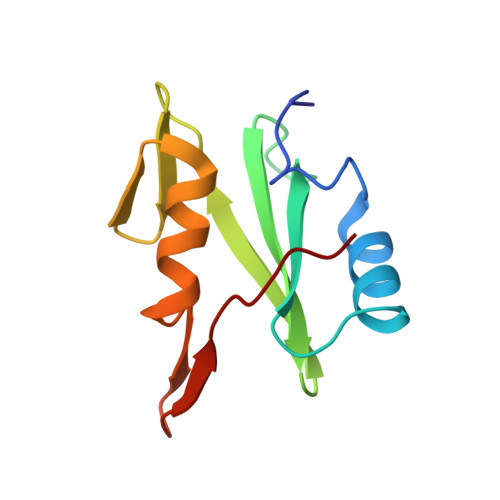
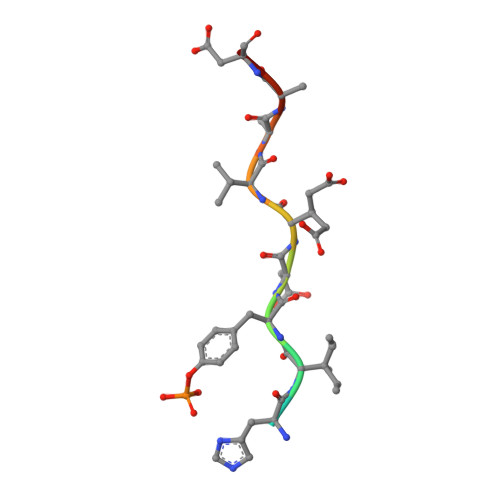
The Phosphotyrosine Peptide Binding Specificity of Nck1 and Nck2 Src Homology 2 Domains.
Frese, S., Schubert, W.-D., Findeis, A.C., Marquardt, T., Roske, Y.S., Stradal, T.E.B., Heinz, D.W.(2006) J Biological Chem 281: 18236
- PubMed: 16636066
- DOI: https://doi.org/10.1074/jbc.M512917200
- Primary Citation of Related Structures:
2CI8, 2CI9, 2CIA - PubMed Abstract:
Nck proteins are essential Src homology (SH) 2 and SH3 domain-bearing adapters that modulate actin cytoskeleton dynamics by linking proline-rich effector molecules to tyrosine kinases or phosphorylated signaling intermediates. Two mammalian pathogens, enteropathogenic Escherichia coli and vaccinia virus, exploit Nck as part of their infection strategy. Conflicting data indicate potential differences in the recognition specificities of the SH2 domains of the isoproteins Nck1 (Nckalpha) and Nck2 (Nckbeta and Grb4). We have characterized the binding specificities of both SH2 domains and find them to be essentially indistinguishable. Crystal structures of both domains in complex with phosphopeptides derived from the enteropathogenic E. coli protein Tir concur in identifying highly conserved, specific recognition of the phosphopeptide. Differential peptide recognition can therefore not account for the preference of either Nck in particular signaling pathways. Binding studies using sequentially mutated, high affinity phosphopeptides establish the sequence variability tolerated in peptide recognition. Based on this binding motif, we identify potential new binding partners of Nck1 and Nck2 and confirm this experimentally for the Arf-GAP GIT1.
- Division of Cell Biology, German Research Center for Biotechnology, Mascheroder Weg 1, D-38124 Braunschweig, Germany.
Organizational Affiliation: