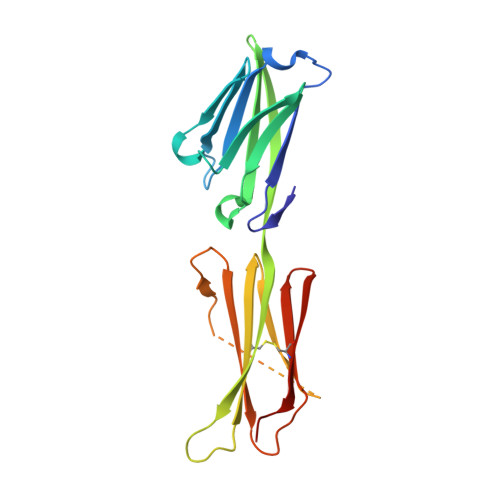
Structure of the Epstein-Barr Virus Oncogene Barf1
Tarbouriech, N., Ruggiero, F., Deturenne-Tessier, M., Ooka, T., Burmeister, W.P.(2006) J Mol Biology 359: 667
- PubMed: 16647084
- DOI: https://doi.org/10.1016/j.jmb.2006.03.056
- Primary Citation of Related Structures:
2CH8 - PubMed Abstract:
The Epstein-Barr virus is a human gamma-herpesvirus that persistently infects more than 90% of the human population. It is associated with numerous epithelial cancers, principally undifferentiated nasopharyngeal carcinoma and gastric carcinoma. The BARF1 gene is expressed in a high proportion of these cancers. An oncogenic, mitogenic and immortalizing activity of the BARF1 protein has been shown. We solved the structure of the secreted BARF1 glycoprotein expressed in a human cell line by X-ray crystallography at a resolution of 2.3A. The BARF1 protein consists of two immunoglobulin (Ig)-like domains. The N-terminal domain belongs to the subfamily of variable domains whereas the C-terminal one is related to a constant Ig-domain. BARF1 shows an unusual hexamerisation involving two principal contacts, one between the C-terminal domains and one between the N-terminal domains. The C-terminal contact with an uncommonly large contact surface extends the beta-sandwich of the Ig-domain through the second molecule. The N-terminal contact involves Ig-domains with an unusual relative orientation but with a more classical contact surface with a size in the range of dimer interactions of Ig-domains. The structure of BARF1 is most closely related to CD80 or B7-1, a co-stimulatory molecule present on antigen presenting cells, from which BARF1 must have been derived during evolution. Still, domain orientation and oligomerization differ between BARF1 and CD80. It had been shown that BARF1 binds to hCSF-1, the human colony-stimulating factor 1, but this interaction has to be principally different from the one between CSF-1 and CSF-1 receptor.
- EMBL Grenoble outstation, BP181, F-38042, Grenoble cedex 9, France.
Organizational Affiliation: