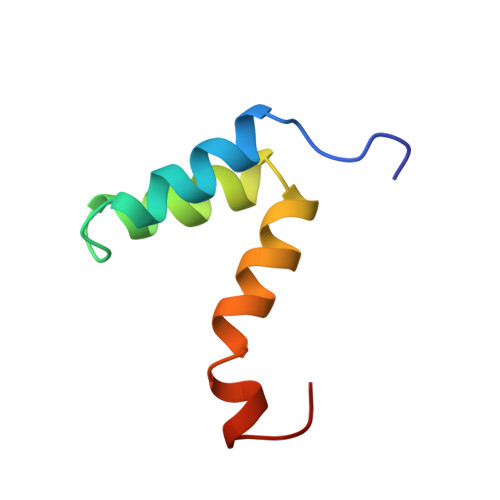
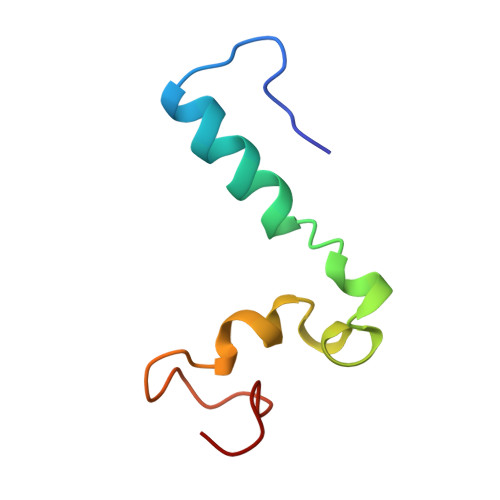
Structural diversity in p160/CREB-binding protein coactivator complexes.
Waters, L., Yue, B., Veverka, V., Renshaw, P., Bramham, J., Matsuda, S., Frenkiel, T., Kelly, G., Muskett, F., Carr, M., Heery, D.M.(2006) J Biological Chem 281: 14787-14795
- PubMed: 16540468
- DOI: https://doi.org/10.1074/jbc.M600237200
- Primary Citation of Related Structures:
2C52 - PubMed Abstract:
Ligand-induced transcription by nuclear receptors involves the recruitment of p160 coactivators such as steroid receptor coactivator 1 (SRC1), in complex with histone acetyltransferases such as CREB-binding protein (CBP) and p300. Here we describe the solution structure of a complex formed by the SRC1 interaction domain (SID) of CBP and the activation domain (AD1) of SRC1, both of which contain four helical regions (Calpha1, Calpha2, Calpha3, and Calpha3' in CBP and Salpha1, Salpha2', Salpha2, and Salpha3 in SRC1). A tight four-helix bundle is formed between Salpha1, Calpha1, Calpha2, and Calpha3 that is capped by Salpha3. In contrast to the structure of the AD1 domain of the related p160 protein ACTR in complex with CBP SID, the sequences forming Salpha2' and Salpha2 in SRC1 AD1 are not involved in the interface between the two domains but rather serve to position Salpha3. Thus, although the CBP SID domain adopts a similar fold in complex with different p160 proteins, the topologies of the AD1 domains are strikingly different, a feature that is likely to contribute to functional specificity of these coactivator complexes.
- Department of Biochemistry, Henry Wellcome Building, University of Leicester, Lancaster Road, Leicester LE1 9HN, United Kingdom.
Organizational Affiliation: