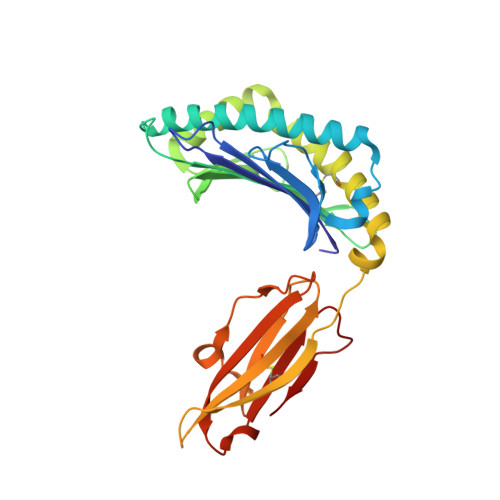
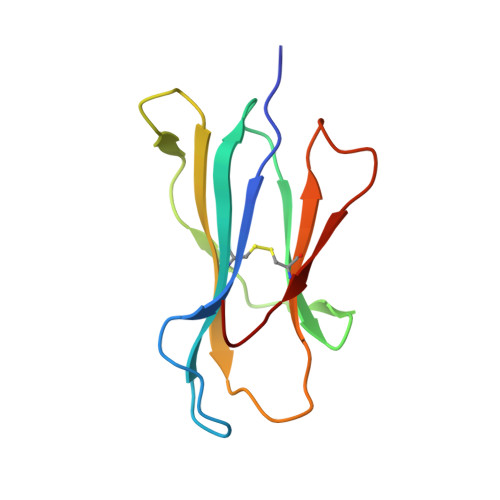
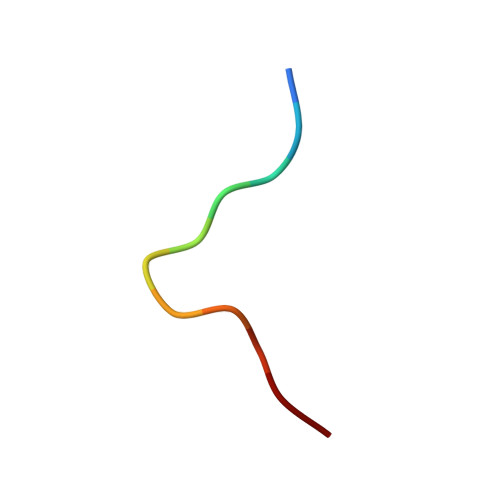Structures of three HIV-1 HLA-B*5703-peptide complexes and identification of related HLAs potentially associated with long-term nonprogression.
Stewart-Jones, G.B., Gillespie, G., Overton, I.M., Kaul, R., Roche, P., McMichael, A.J., Rowland-Jones, S., Jones, E.Y.(2005) J Immunol 175: 2459-2468
- PubMed: 16081817
- DOI: https://doi.org/10.4049/jimmunol.175.4.2459
- Primary Citation of Related Structures:
2BVO, 2BVP, 2BVQ - PubMed Abstract:
Long-term nonprogression during acute HIV infection has been strongly associated with HLA-B*5701 or HLA-B*5703. In this study, we present the high resolution crystal structures of HLA-B*5703 complexes with three HIV-1 epitopes: ISPRTLNAW (ISP), KAFSPEVIPMF (KAF-11), and KAFSPEVI (KAF-8). These reveal peptide anchoring at position 2 and their C termini. The different peptide lengths and primary sequences are accommodated by variation in the specific contacts made to the HLA-B*5703, flexibility in water structure, and conformational adjustment of side chains within the peptide-binding groove. The peptides adopt markedly different conformations, and trap variable numbers of water molecules, near a cluster of tyrosine side chains located in the central region of the peptide-binding groove. The KAF-11 epitope completely encompasses the shorter KAF-8 epitope but the peptides are presented in different conformations; the KAF-11 peptide arches out of the peptide-binding groove, exposing a significant main chain surface area. Bioinformatic analysis of the MHC side chains observed to contribute to the peptide anchor specificity, and other specific peptide contacts, reveals HLA alleles associated with long-term nonprogression and a number of related HLA alleles that may share overlapping peptide repertoires with HLA-B*5703 and thus may display a similar capacity for efficient immune control of HIV-1 infection.
- Division of Structural Biology, Wellcome Trust Centre for Human Genetics, University of Oxford, Oxford, United Kingdom.
Organizational Affiliation: