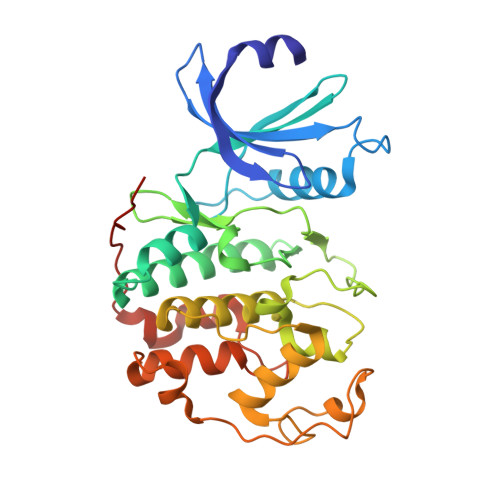
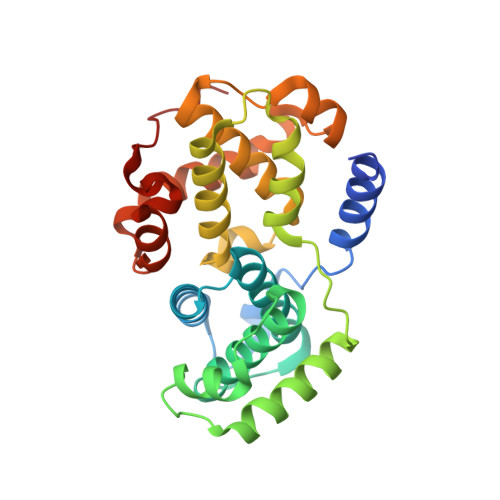
Benzodipyrazoles: A New Class of Potent Cdk2 Inhibitors
Dalessio, R., Bargiottia, A., Metz, S., Brasca, M.G., Cameron, A., Ermoli, A., Marsiglio, A., Polucci, P., Roletto, F., Tibolla, M., Vazquez, M.L., Vulpetti, A., Pevarello, P.(2005) Bioorg Med Chem Lett 15: 1315
- PubMed: 15713378
- DOI: https://doi.org/10.1016/j.bmcl.2005.01.023
- Primary Citation of Related Structures:
2BKZ - PubMed Abstract:
The synthesis and the preliminary expansion of this new class of CDK2 inhibitors are presented. The synthesis was accomplished using a solution-phase protocol amenable to rapid parallel expansion and suitable to be scaled-up in view of possible lead development. Following a medicinal chemistry program aimed at improving cell permeability and selectivity, a series of compounds with nanomolar activity in the biochemical assay and able to efficiently inhibit tumor cell proliferation has been obtained.
- Chemistry Department, Nerviano Medical Sciences, Oncology Business Unit, Viale Pasteur 10, 20014 Nerviano (MI), Italy. roberto.dalessio@nervianoms.com
Organizational Affiliation: