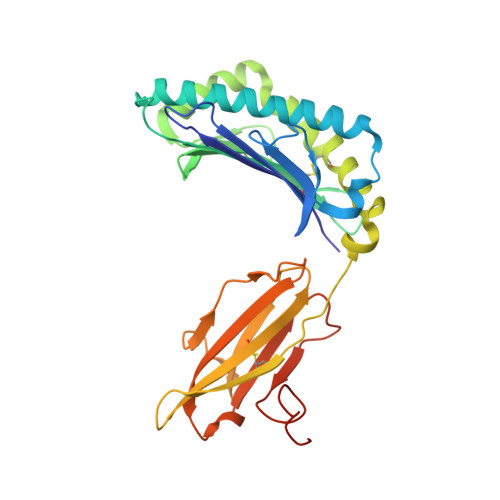
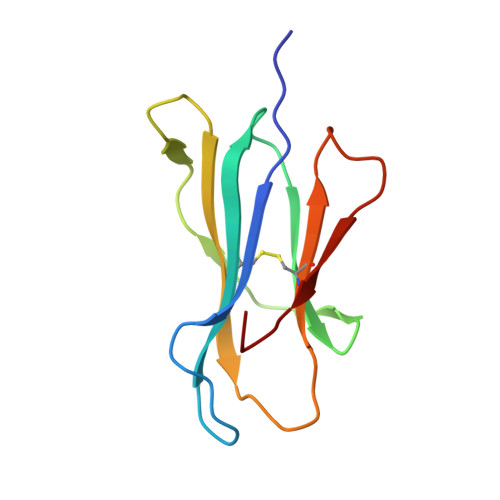
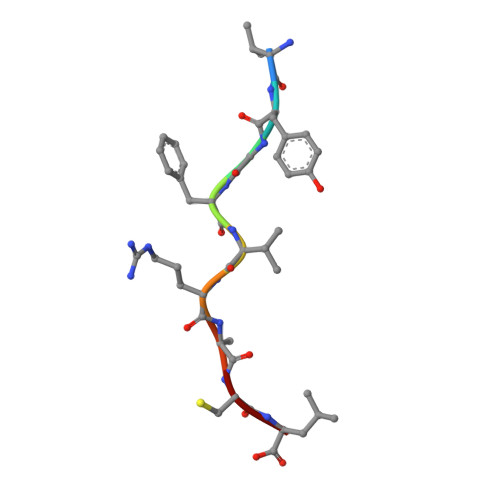
Crystal structure of HLA-A*2402 complexed with a telomerase peptide.
Cole, D.K., Rizkallah, P.J., Gao, F., Watson, N.I., Boulter, J.M., Bell, J.I., Sami, M., Gao, G.F., Jakobsen, B.K.(2006) Eur J Immunol 36: 170-179
- PubMed: 16323248
- DOI: https://doi.org/10.1002/eji.200535424
- Primary Citation of Related Structures:
2BCK - PubMed Abstract:
HLA-A*2402 is the most commonly expressed HLA allele in oriental populations. It is also widely expressed in the Caucasian population, making it one of, if not the most abundant HLA I types. In order to study its structure in terms of overall fold and peptide presentation, a soluble form of this HLA I (alpha1, alpha2, alpha3 and beta(2)m domains) has been expressed, refolded and crystallized in complex with a cancer-related telomerase peptide (VYGFVRACL), and its structure has been solved to 2.8 A resolution. The overall structure of HLA-A*2402 is virtually identical to other reported peptide-HLA I structures. However, there are distinct features observable from this structure at the HLA I peptide binding pockets. The size and depth of pocket B makes it highly suitable for binding to large aromatic side chains, which explains the high prevalence of tyrosine at peptide position 2. Also, for HLA binding at peptide position 5, there is an additional anchor point, which allows the proximal amino acids to protrude out, providing a prominent feature for TCR interaction. Finally, pocket F allows the anchor residue at position 9 to be bound unusually deeply within the HLA structure.
- Nuffield Dept. of Clinical Medicine, John Radcliffe Hospital, Oxford University, Oxford, UK.
Organizational Affiliation: