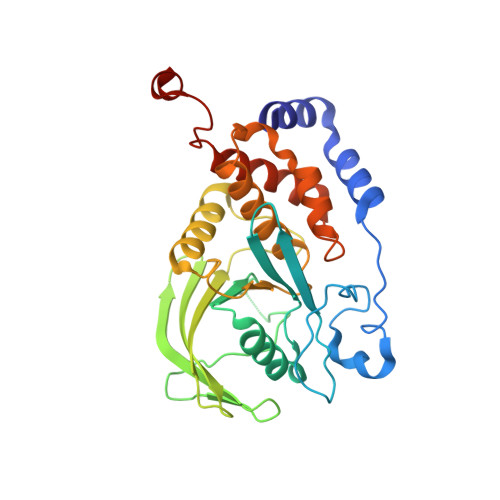
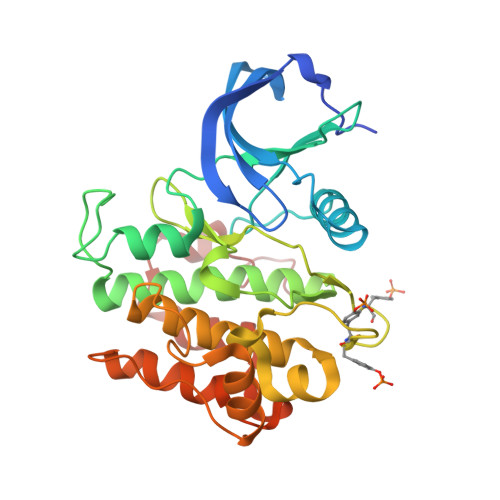
Crystal Structure of a Complex between Protein Tyrosine Phosphatase 1B and the Insulin Receptor Tyrosine Kinase.
Li, S., Depetris, R.S., Barford, D., Chernoff, J., Hubbard, S.R.(2005) Structure 13: 1643-1651
- PubMed: 16271887
- DOI: https://doi.org/10.1016/j.str.2005.07.019
- Primary Citation of Related Structures:
2B4S - PubMed Abstract:
Protein tyrosine phosphatase 1B (PTP1B) is a highly specific negative regulator of insulin receptor signaling in vivo. The determinants of PTP1B specificity for the insulin receptor versus other receptor tyrosine kinases are largely unknown. Here, we report a crystal structure at 2.3 A resolution of the catalytic domain of PTP1B (trapping mutant) in complex with the phosphorylated tyrosine kinase domain of the insulin receptor (IRK). The crystallographic asymmetric unit contains two PTP1B-IRK complexes that interact through an IRK dimer interface. Rather than binding to a phosphotyrosine in the IRK activation loop, PTP1B binds instead to the opposite side of the kinase domain, with the phosphorylated activation loops sequestered within the IRK dimer. The crystal structure provides evidence for a noncatalytic mode of interaction between PTP1B and IRK, which could be important for the selective recruitment of PTP1B to the insulin receptor.
- Structural Biology Program, Skirball Institute of Biomolecular Medicine and Department of Pharmacology, New York University School of Medicine, New York, New York 10016, USA.
Organizational Affiliation: