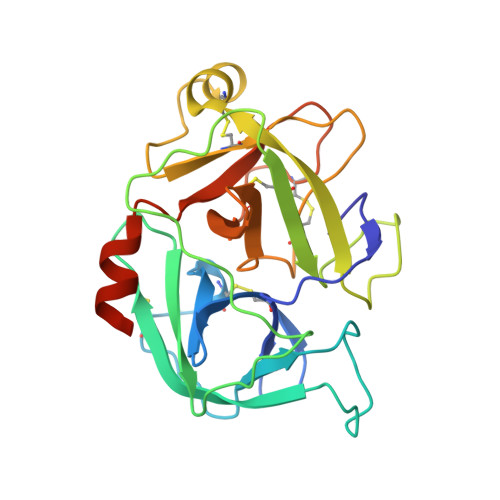
Crystal structure of the beta-chain of human hepatocyte growth factor-like/macrophage stimulating protein.
Carafoli, F., Chirgadze, D.Y., Blundell, T.L., Gherardi, E.(2005) FEBS J 272: 5799-5807
- PubMed: 16279944
- DOI: https://doi.org/10.1111/j.1742-4658.2005.04968.x
- Primary Citation of Related Structures:
2ASU - PubMed Abstract:
Hepatocyte growth factor like/macrophage stimulating protein (HGFl/MSP) and hepatocyte growth factor/scatter factor (HGF/SF) define a distinct family of vertebrate-specific growth factors structurally related to the blood proteinase precursor plasminogen and with important roles in development and cancer. Although the two proteins share a similar domain structure and mechanism of activation, there are differences between HGFl/MSP and HGF/SF in terms of the contribution of individual domains to receptor binding. Here we present a crystal structure of the 30 kDa beta-chain of human HGFl/MSP, a serine proteinase homology domain containing the high-affinity binding site for the RON receptor. The structure describes at 1.85 Angstrom resolution the region of the domain corresponding to the receptor binding site recently defined in the HGF/SF beta-chain, namely the central cleft harboring the three residues corresponding to the catalytic ones of active proteinases (numbers in brackets define the sequence position according to the standard chymotrypsinogen numbering system) [Gln522 (c57), Gln568 (c102) and Tyr661 (c195)] and an adjacent loop flanking the S1 specificity pocket and containing residues Asn682 (c217) and Arg683 (c218) previously shown to be essential for binding of HGFl/MSP to the RON receptor. The study confirms the concept that the serine proteinase homology domains of HGFl/MSP and HGF/SF bind their receptors in an 'enzyme-substrate' mode, reflecting the common evolutionary origin of the plasminogen-related growth factors and the proteinases of the clotting and fibrinolytic pathways. However, analysis of the intermolecular interactions in the crystal lattice of beta-chain HGFl/MSP fails to show the same contacts seen in the HGF/SF structures and does not support a conserved mode of dimerization of the serine proteinase homology domains of HGFl/MSP and HGF/SF responsible for receptor activation.
- MRC Centre, Hills Road, Cambridge, UK.
Organizational Affiliation: