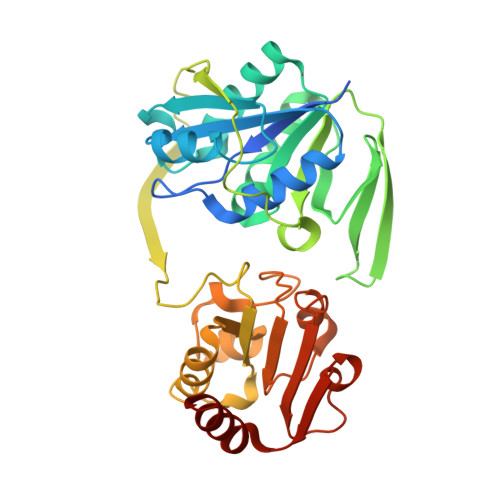
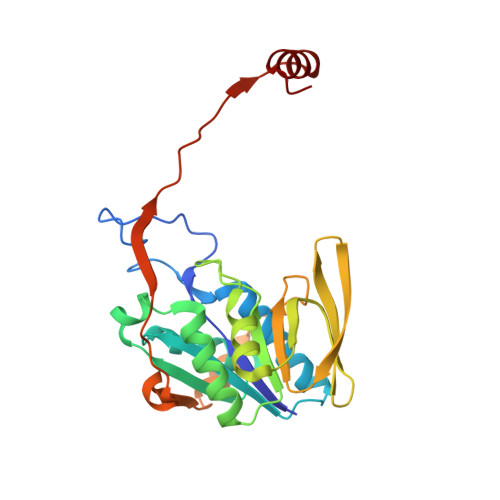
Stabilization of Non-productive Conformations Underpins Rapid Electron Transfer to Electron-transferring Flavoprotein
Toogood, H.S., van Thiel, A., Scrutton, N.S., Leys, D.(2005) J Biological Chem 280: 30361-30366
- PubMed: 15975918
- DOI: https://doi.org/10.1074/jbc.M505562200
- Primary Citation of Related Structures:
2A1T, 2A1U - PubMed Abstract:
Crystal structures of protein complexes with electron-transferring flavoprotein (ETF) have revealed a dual protein-protein interface with one region serving as anchor while the ETF FAD domain samples available space within the complex. We show that mutation of the conserved Glu-165beta in human ETF leads to drastically modulated rates of interprotein electron transfer with both medium chain acyl-CoA dehydrogenase and dimethylglycine dehydrogenase. The crystal structure of free E165betaA ETF is essentially identical to that of wild-type ETF, but the crystal structure of the E165betaA ETF.medium chain acyl-CoA dehydrogenase complex reveals clear electron density for the FAD domain in a position optimal for fast interprotein electron transfer. Based on our observations, we present a dynamic multistate model for conformational sampling that for the wild-type ETF. medium chain acyl-CoA dehydrogenase complex involves random motion between three distinct positions for the ETF FAD domain. ETF Glu-165beta plays a key role in stabilizing positions incompatible with fast interprotein electron transfer, thus ensuring high rates of complex dissociation.
- Department of Biochemistry, University of Leicester, Henry Wellcome Building, Lancaster Road, LE1 7RH, Leicester United Kingdom.
Organizational Affiliation: