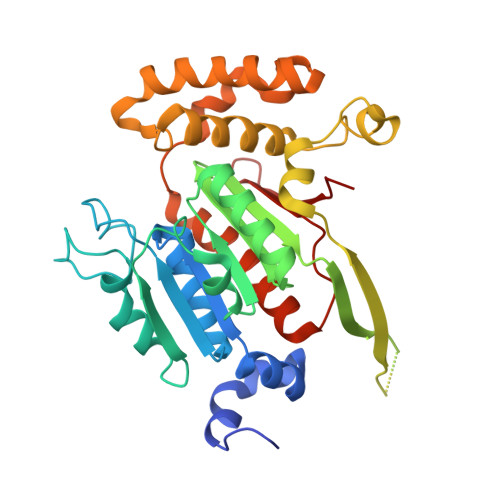
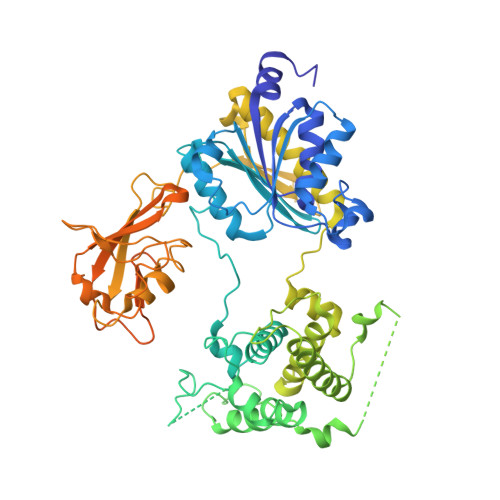
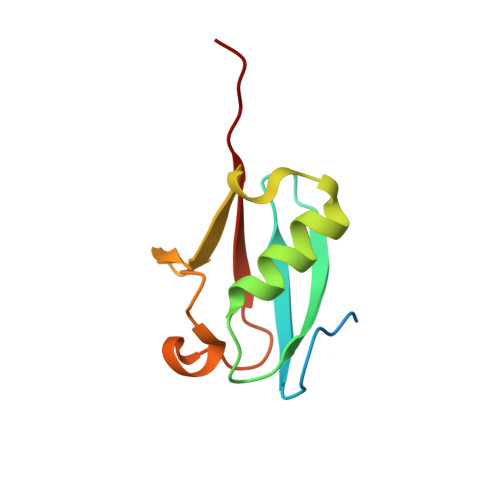
Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1
Lois, L.M., Lima, C.D.(2005) EMBO J 24: 439-451
- PubMed: 15660128
- DOI: https://doi.org/10.1038/sj.emboj.7600552
- Primary Citation of Related Structures:
1Y8Q, 1Y8R - PubMed Abstract:
E1 enzymes facilitate conjugation of ubiquitin and ubiquitin-like proteins through adenylation, thioester transfer within E1, and thioester transfer from E1 to E2 conjugating proteins. Structures of human heterodimeric Sae1/Sae2-Mg.ATP and Sae1/Sae2-SUMO-1-Mg.ATP complexes were determined at 2.2 and 2.75 A resolution, respectively. Despite the presence of Mg.ATP, the Sae1/Sae2-SUMO-1-Mg.ATP structure reveals a substrate complex insomuch as the SUMO C-terminus remains unmodified within the adenylation site and 35 A from the catalytic cysteine, suggesting that additional changes within the adenylation site may be required to facilitate chemistry prior to adenylation and thioester transfer. A mechanism for E2 recruitment to E1 is suggested by biochemical and genetic data, each of which supports a direct role for the E1 C-terminal ubiquitin-like domain for E2 recruitment during conjugation.
- Structural Biology Program, Sloan-Kettering Institute, New York, NY 10021, USA.
Organizational Affiliation: