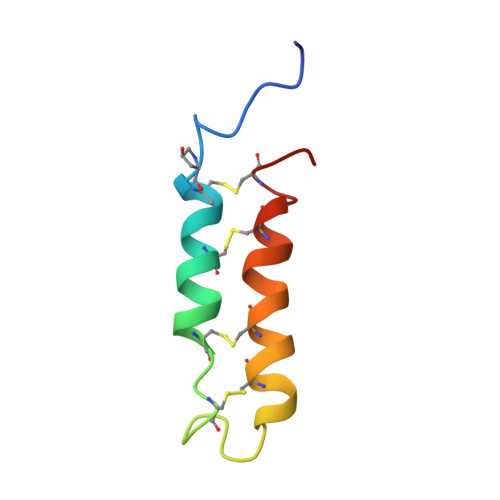
Structure of neurotoxin B-IV from the marine worm Cerebratulus lacteus: a helical hairpin cross-linked by disulphide bonding.
Barnham, K.J., Dyke, T.R., Kem, W.R., Norton, R.S.(1997) J Mol Biology 268: 886-902
- PubMed: 9180379
- DOI: https://doi.org/10.1006/jmbi.1997.0980
- Primary Citation of Related Structures:
1VIB - PubMed Abstract:
B-IV is a 55-residue, crustacean-selective, neurotoxin secreted by Cerebratulus lacteus, a large marine worm found along the northeastern coast of North America. The 3D structure of this molecule in aqueous solution has been determined by 1H NMR spectroscopy at 600 MHz. The molecule has a well-defined helical hairpin structure, with the branches of the hairpin linked by four disulphide bonds. The disulphide connectivities were established from the NMR data to be 1-8/2-7/3-6/4-5, which differed from those determined previously by chemical means, where 1-7 and 2-8 connectivities were found. Each branch of the hairpin is largely alpha-helical, with the helices in the N and C-terminal branches encompassing residues 11 to 23 and 34 to 49, respectively. The loop connecting the branches of the hairpin contains two inverse gamma-turns centred on residues 24 and 25, a type I beta-turn at residues 28 to 31 and a type II beta-turn at residues 30 to 33. Arg17, -25 and -34, which are important for activity, are all on the same face of the molecule, while Trp30, which is also important for activity, is on the opposite face. Structure comparisons show that the B-IV structure is quite similar to those of Rop (ColE1 repressor of primer) and the heat-stable enterotoxin B from Escherichia coli. These structural similarities are discussed in relation to possible mechanisms of action of B-IV.
- Biomolecular Research Institute, Parkville, Victoria, Australia.
Organizational Affiliation: