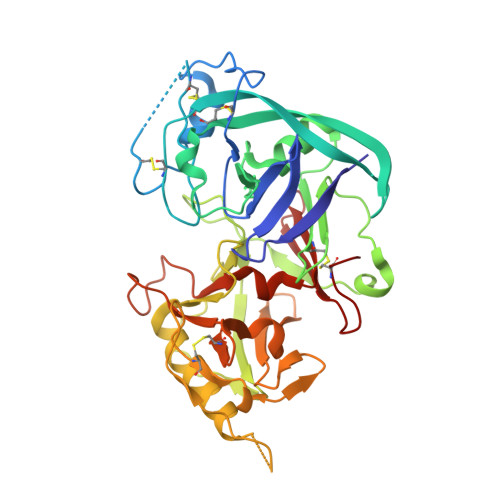
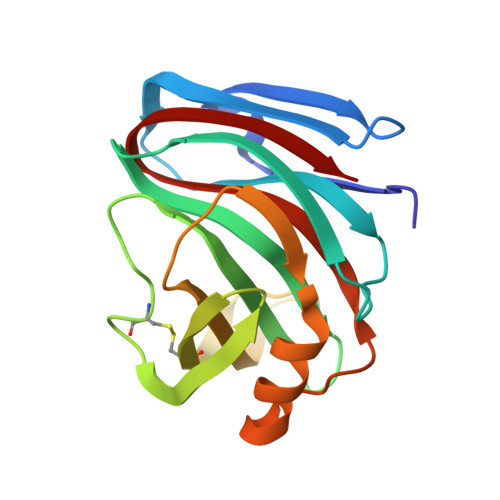
Structural basis for inhibition of Aspergillus niger xylanase by triticum aestivum xylanase inhibitor-I
Sansen, S., De Ranter, C.J., Gebruers, K., Brijs, K., Courtin, C.M., Delcour, J.A., Rabijns, A.(2004) J Biological Chem 279: 36022-36028
- PubMed: 15166216
- DOI: https://doi.org/10.1074/jbc.M404212200
- Primary Citation of Related Structures:
1T6E, 1T6G - PubMed Abstract:
Plants developed a diverse battery of defense mechanisms in response to continual challenges by a broad spectrum of pathogenic microorganisms. Their defense arsenal includes inhibitors of cell wall-degrading enzymes, which hinder a possible invasion and colonization by antagonists. The structure of Triticum aestivum xylanase inhibitor-I (TAXI-I), a first member of potent TAXI-type inhibitors of fungal and bacterial family 11 xylanases, has been determined to 1.7-A resolution. Surprisingly, TAXI-I displays structural homology with the pepsin-like family of aspartic proteases but is proteolytically nonfunctional, because one or more residues of the essential catalytical triad are absent. The structure of the TAXI-I. Aspergillus niger xylanase I complex, at a resolution of 1.8 A, illustrates the ability of tight binding and inhibition with subnanomolar affinity and indicates the importance of the C-terminal end for the differences in xylanase specificity among different TAXI-type inhibitors.
- Laboratory of Analytical Chemistry and Medicinal Physicochemistry, Katholieke Universiteit Leuven, E. van Evenstraat 4, B-3000 Leuven, Belgium.
Organizational Affiliation: