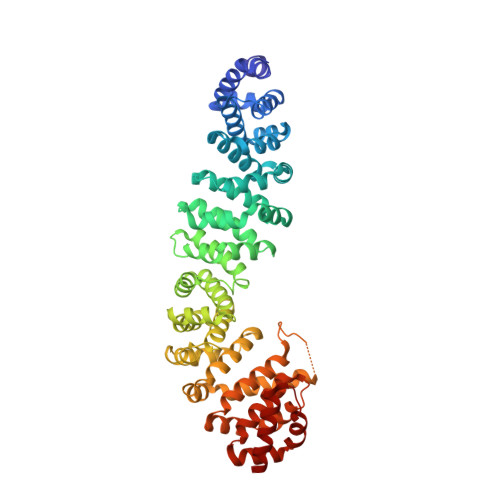
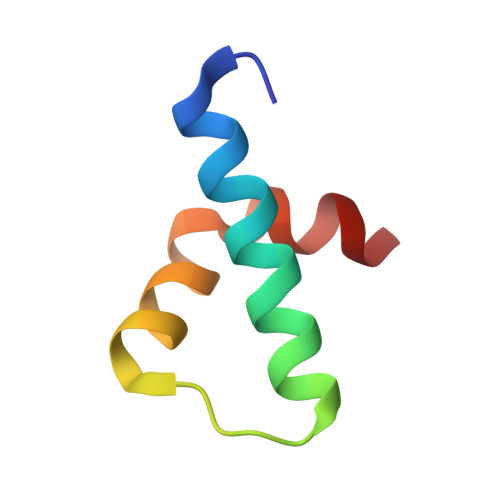
Mechanism of phosphorylation-dependent binding of APC to beta-catenin and its role in beta-catenin degradation
Ha, N.-C., Tonozuka, T., Stamos, J.L., Choi, H.J., Weis, W.I.(2004) Mol Cell 15: 511-521
- PubMed: 15327768
- DOI: https://doi.org/10.1016/j.molcel.2004.08.010
- Primary Citation of Related Structures:
1T08, 1V18 - PubMed Abstract:
The transcriptional coactivator beta-catenin mediates Wnt growth factor signaling. In the absence of a Wnt signal, casein kinase 1 (CK1) and glycogen synthase kinase-3beta (GSK-3beta) phosphorylate cytosolic beta-catenin, thereby flagging it for recognition and destruction by the ubiquitin/proteosome machinery. Phosphorylation occurs in a multiprotein complex that includes the kinases, beta-catenin, axin, and the Adenomatous Polyposis Coli (APC) protein. The role of APC in this process is poorly understood. CK1epsilon and GSK-3beta phosphorylate APC, which increases its affinity for beta-catenin. Crystal structures of phosphorylated and nonphosphorylated APC bound to beta-catenin reveal a phosphorylation-dependent binding motif generated by mutual priming of CK1 and GSK-3beta substrate sequences. Axin is shown to act as a scaffold for substrate phosphorylation by these kinases. Phosphorylated APC and axin bind to the same surface of, and compete directly for, beta-catenin. The structural and biochemical data suggest a novel model for how APC functions in beta-catenin degradation.
- Department of Structural Biology, Stanford University School of Medicine, Stanford, CA 94043, USA.
Organizational Affiliation: