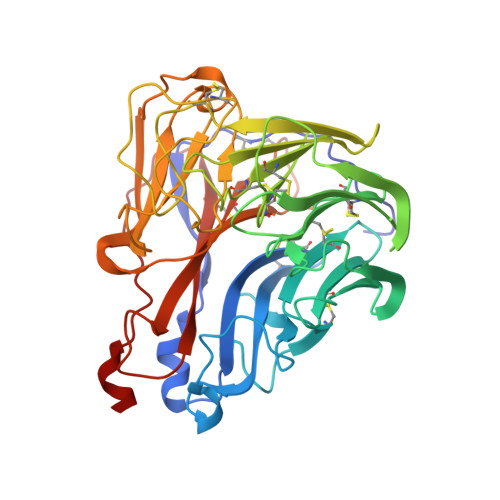
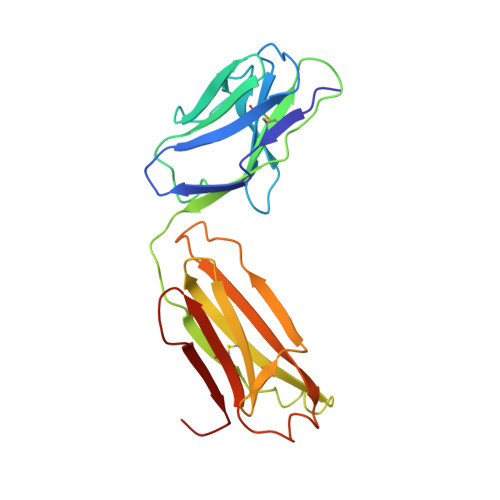
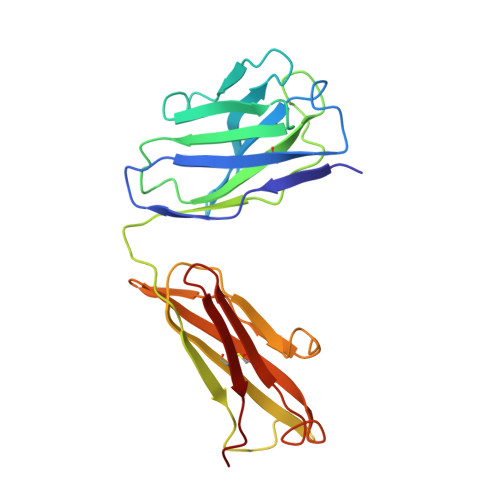
Refined crystal structure of the influenza virus N9 neuraminidase-NC41 Fab complex.
Tulip, W.R., Varghese, J.N., Laver, W.G., Webster, R.G., Colman, P.M.(1992) J Mol Biology 227: 122-148
- PubMed: 1381757
- DOI: https://doi.org/10.1016/0022-2836(92)90687-f
- Primary Citation of Related Structures:
1NCA, 1NCD - PubMed Abstract:
The crystal structure of the complex between neuraminidase from influenza virus (subtype N9 and isolated from an avian source) and the antigen-binding fragment (Fab) of monoclonal antibody NC41 has been refined by both least-squares and simulated annealing methods to an R-factor of 0.191 using 31,846 diffraction data in the resolution range 8.0 to 2.5 A. The resulting model has a root-mean-square deviation from ideal bond-length of 0.016 A. One fourth of the tetrameric complex comprises the crystallographic model, which has 6577 non-hydrogen atoms and consists of 389 protein residues and eight carbohydrate residues in the neuraminidase, 214 residues in the Fab light chain, and 221 residues in the heavy chain. One putative Ca ion buried in the neuraminidase, and 73 water molecules, are also included. A remarkable shape complementarity exists between the interacting surfaces of the antigen and the antibody, although the packing density of atoms at the interface is somewhat looser than in the interior of a protein. Similarly, there is a high degree of chemical complementarity between the antigen and antibody, mediated by one buried salt-link, two solvated salt-links and 12 hydrogen bonds. The antibody-binding site on neuraminidase is discontinuous and comprises five chain segments and 19 residues in contact, whilst 33 neuraminidase residues in eight segments have 899 A2 of surface area buried by the interaction (to a 1.7 A probe), including two hexose units. Seventeen residues in NC41 Fab lying in five of the six complementarity determining regions (CDRs) make contact with the neuraminidase and 36 antibody residues in seven segments have 916 A2 of buried surface area. The interface is more extensive than those of the three lysozyme-Fab complexes whose crystal structures have been determined, as judged by buried surface area and numbers of contact residues. There are only small differences (less than 1.5 A) between the complexed and uncomplexed neuraminidase structures and, at this resolution and accuracy, those differences are not unequivocal. The main-chain conformations of five of the CDRs follow the predicted canonical structures. The interface between the variable domains of the light and heavy chains is not as extensive as in other Fabs, due to less CDR-CDR interaction in NC41. The first CDR on the NC41 Fab light chain is positioned so that it could sterically hinder the approach of small as well as large substrates to the neuraminidase active-site pocket, suggesting a possible mechanism for the observed inhibition of enzyme activity by the antibody.(ABSTRACT TRUNCATED AT 400 WORDS)
- CSIRO Division of Biomolecular Engineering, Parkville, Victoria, Australia.
Organizational Affiliation: