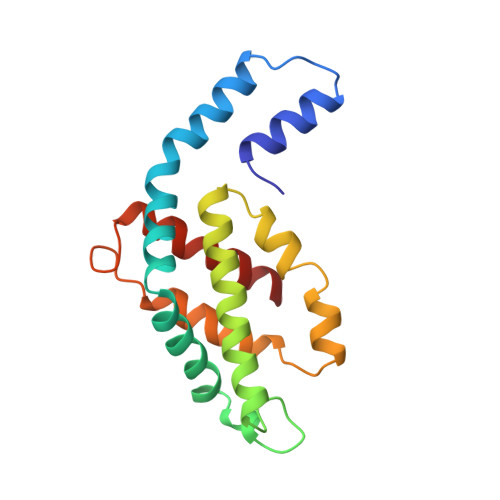
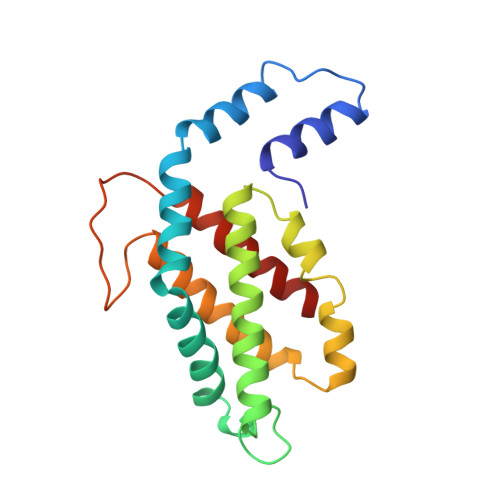
Crystal structure of R-phycoerythrin from Polysiphonia urceolata at 2.8 A resolution.
Chang, W.R., Jiang, T., Wan, Z.L., Zhang, J.P., Yang, Z.X., Liang, D.C.(1996) J Mol Biology 262: 721-731
- PubMed: 8876649
- DOI: https://doi.org/10.1006/jmbi.1996.0547
- Primary Citation of Related Structures:
1LIA - PubMed Abstract:
The structure of R-phycoerythrin (R-PE) from Polysiphonia urceolata was determined at 2.8 A resolution. The crystals belong to space group R3 with unit cell dimensions of a = b = 189.8 A, c = 60.1 A. The subunit composition of R-PE is (alpha 2 beta 2)3 gamma. The three-dimensional structure of R-PE was solved by the multiple isomorphous replacement method. After several cycles of model building and refinement, the crystallographic R-factor of the final model is 18.0% with data from 10.0 to 2.8 A resolution. The four phycoerythrobilin chromophores alpha 84, alpha 140a, beta 84 and beta 155 in an (alpha beta) unit are each covalently bound to a cysteine residue through ring A. The phycourobilin chromophore is bound to cysteine beta 50 by ring A and bound to cysteine beta 61 by ring D. The ring A and ring D of phycourobilin deviate from the conjugate plane formed by ring B and ring C and the four rings form a boat-shaped structure. R-PE contains a 34 kDa gamma subunit that is assumed to lie in the central channel of the molecular disc (alpha 2 beta 2)3. The energy transfer and relationship between cysteine residues and chromophores are discussed.
- National Laboratory of Biomacromolecules, Chinese Academy of Sciences, Beijing, China.
Organizational Affiliation: