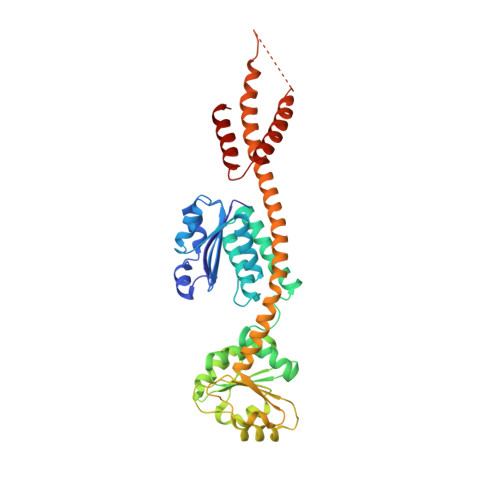
V-shaped structure of glutamyl-tRNA reductase, the first enzyme of tRNA-dependent tetrapyrrole biosynthesis.
Moser, J., Schubert, W.D., Beier, V., Bringemeier, I., Jahn, D., Heinz, D.W.(2001) EMBO J 20: 6583-6590
- PubMed: 11726494
- DOI: https://doi.org/10.1093/emboj/20.23.6583
- Primary Citation of Related Structures:
1GPJ - PubMed Abstract:
Processes vital to life such as respiration and photosynthesis critically depend on the availability of tetrapyrroles including hemes and chlorophylls. tRNA-dependent catalysis generally is associated with protein biosynthesis. An exception is the reduction of glutamyl-tRNA to glutamate-1-semialdehyde by the enzyme glutamyl-tRNA reductase. This reaction is the indispensable initiating step of tetrapyrrole biosynthesis in plants and most prokaryotes. The crystal structure of glutamyl-tRNA reductase from the archaeon Methanopyrus kandleri in complex with the substrate-like inhibitor glutamycin at 1.9 A resolution reveals an extended yet planar V-shaped dimer. The well defined interactions of the inhibitor with the active site support a thioester-mediated reduction process. Modeling the glutamyl-tRNA onto each monomer reveals an extensive protein-tRNA interface. We furthermore propose a model whereby the large void of glutamyl-tRNA reductase is occupied by glutamate-1-semialdehyde-1,2-mutase, the subsequent enzyme of this pathway, allowing for the efficient synthesis of 5-aminolevulinic acid, the common precursor of all tetrapyrroles.
- Institute of Microbiology, Technical University Braunschweig, Spielmannstrasse 7, D-38106 Braunschweig, Germany.
Organizational Affiliation: